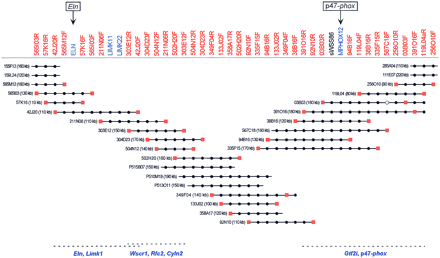
BAC/PAC-based STS-content map of the mouse WS region (oriented with centromere leftward and telomere rightward). The deduced positions of 44 STSs are depicted along the top, with the indicated clones shown as horizontal lines below. Relevant information about the STSs is available in GenBank (http://www.ncbi.nlm.nih.gov). The STSs were developed from clone insert ends (red), known genes (blue), or a conserved human DNA sequence (black). (●,◼) The STS is confirmed to be present in that clone by PCR testing. When an STS corresponds to a clone insert end, a red square is present at the end of the clone from which it was derived. PAC clones are indicated with a P at the beginning of their names. Four of the BAC clones were isolated from the Genome Systems C57BL/6 mouse library (111E07, 285A04, 155F12, and 159L24), with the rest of the BACs coming from the Research Genetics CITB–CJ7-B mouse library. The size of each BAC/PAC, as assessed by pulsed-field gel analysis, is also provided. The indicated BAC/PAC overlaps were confirmed by restriction enzyme digest-based fingerprint analysis (Marra et al. 1997; data not shown). The depicted clones together span ∼680 kb of DNA from mouse chromosome 5. Preliminary genomic sequence analysis has revealed the presence ofEln, Limk1, and part of Wscr1 in BAC 42J20,Wscr1, Rfc2, and Cyln2 in BAC 303E12, andGtf2i and p47–phox in BAC 391O16, as indicated along the bottom (U. DeSilva and E.D. Green, unpubl.).











