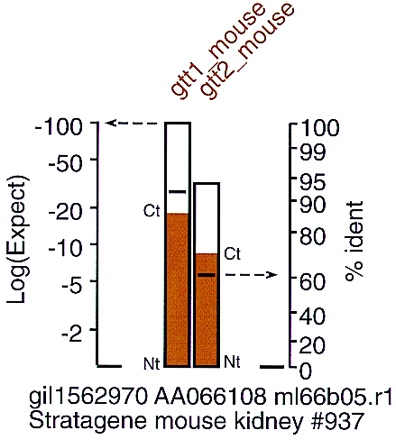
Displaying similarity and identity. FAST_PAN summarizes the similarity of the query protein sequence with the DNA library sequence by plotting a bar with its height determined by the expectation value of the alignment score [left ordinate, log(Expect)]. The log(Expect) is plotted on a logarithmic scale to expand the log(Expect) resolution between −2 and −50. Alignments with expectation values better than 10−50 are typically well recognized already. The horizontal line within the bar indicates the percent identity of the alignment; percent identity is more informative when searching for new subfamily paralogs. The identity axis uses a nonlinear scale to expand the region from 70%–100%. The colored shading in the box is used both to group known families visually and to identify the boundaries of the DNA alignment on the protein query sequence. The bottom of the bar (Nt) indicates the location of the beginning (amino-terminal) of the alignment; the top of the bar (Ct) indicates the end (carboxy-terminal) of the alignment. In the example shown here, the EST sequence contains the amino-terminal two-thirds of the protein query sequences.











