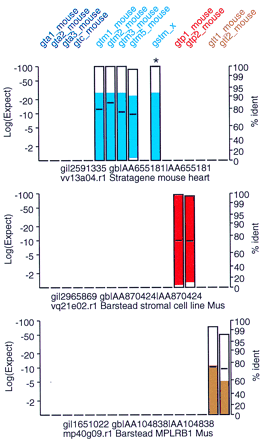
New glutathione transferase paralogs. The mouse EST database used in Fig. 3 was searched with mouse class-alpha, class-mu, class-pi, and class-theta protein sequences. Searches were performed with thetfasty3 program, using the MDM20 (Jones et al. 1992) substitution matrix. Gap penalties were set as in Fig. 3. Included in the plot, but not in the original search, is gstm_x, a novel mouse class-mu glutathione transferase that was assembled from EST sequences sharing less than 90% identity with any of the four known mouse class-mu GSTs. For this example the FAST_PAN program was set to not display sequences sharing 95% identity (or better) with any of the query sequences, except for gstm_x, which is highlighted as >95% identical with an asterisk (*).











