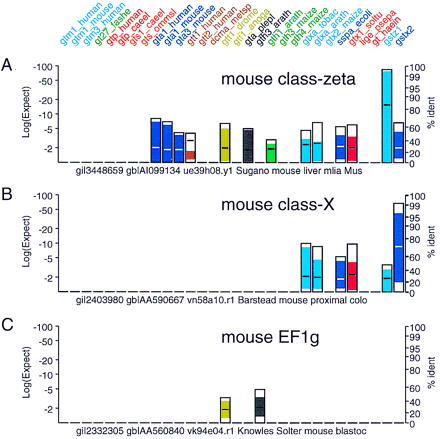
Novel glutathione transferase families. Mouse EST sequences that do not belong to the class-mu, -alpha, -pi, or -theta families are shown. These EST sequences share strong similarity to plant and bacterial glutathione transferases. (Two additional protein query sequences, a mouse class-zeta gst_z and a mouse class-Xgst_x have been added to the panel to highlight the similarity of the novel ESTs with these new glutathione transferase classes.) Searches were performed as in Fig. 3. (A) EST (gi‖3448659) belongs to a recently discovered glutathione transferase family termed class zeta (Board et al. 1997). (B) EST gi‖2403980 (mouse class X), does not share significant similarity with any of the vertebrate sequences in the query set; strongest similarity is seen with theGTXA_TOBAC [E() < 10−10] andGTXA_ARATH [E() < 10−8] glutathione transferases. This EST belongs to a recently described novel mammalian glutathione transferase class (GenBank accession no. U80819, R. Kodym and M.D. Story, unpubl.). (C) EST gi‖2332305shares strong similarity with the EF1γ translation elongation factors and probably does not encode a glutathione transferase. The EF1γ proteins contain a domain homologous to glutathione transferases (Koonin et al. 1994).











