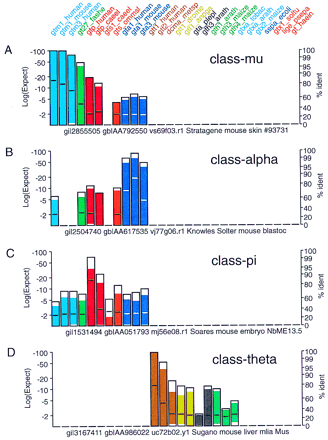
Known glutathione transferase family members. Typical profiles for matches between ESTs encoding members the four well-known mammalian glutathione transferase families: class alpha, class mu, class pi, and class theta. Searches were performed with the glutathione transferase query sequences indicated at the top of the Figure against the est_mouse database obtained from the National Center for Biotechnology Information (http://ncbi.nlm.nih.gov/blast/db/est_mouse) using the tfasty3 program with the default scoring parameters: the BLOSUM50 matrix and −15 for the first residue in a gap, −3 for additional residues, and −20 for frameshifts between or within codons (−15/−3/−20/−20). Matches against class-mu (gi‖2855505), class-alpha (gi‖2504740), class-pi (gi‖1531494), and class-theta (gi‖3167411) ESTs are shown.











