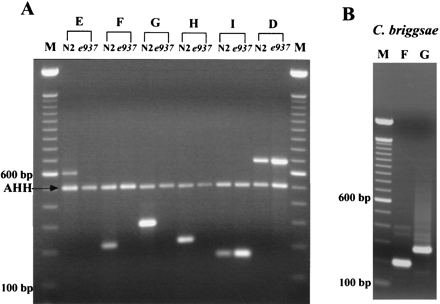
RT–PCR analysis. (A) Examination of bli-4 E, F, G, H, and I expression in C. elegans N2 and e937animals. Amplification was performed using a primer that annealed to exon 12 in combination with a primer specific for each newly predicted exon and bli-4D which resides outside of the e937deletion. Products were amplified for each predicted exon from N2 template RNA, but were missing for exons E (617 bp), F (219 bp), G (300 bp), and H (224 bp) using e937 RNA. The 517-bp product seen in all lanes is the result of amplification using primers designed from the gene encoding S-adenosyl homocysteine hydrolase, used as a positive control. (B) Examination of Cb–bli-4F andbli-4G expression. RNA from C. briggsae animals was used for RT–PCR analysis to determine whether poorly conserved putative F exon was expressed in this species. As can be seen in lane F, amplification using a common region primer in combination with the F specific primer yielded a product of the expected size (199 bp). As a control, we were also able to demonstrate expression of the highly conserved G exon by amplifying the expected 272-bp product (lane G).











