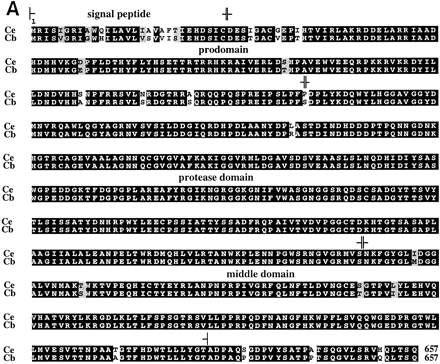
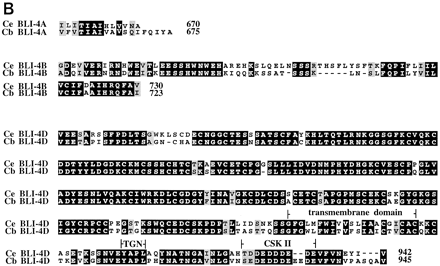
(A) Alignment of the predicted BLI-4 protein sequence encoded by the common region of C. elegans (Ce) and C. briggsae (Cb). Identical residues have been boxed; conserved amino acids are shaded. Residues that are predicted to compose the signal peptide, prodomain, protease domain, and middle domains are shown within the corresponding brackets. (B) Alignment of the predicted carboxyl termini of the conserved BLI-4 isoforms from C. elegans (Ce) and C. briggsae (Cb). Identical residues are boxed; conserved residues are shaded. Conserved amino acid motifs within the carboxyl termini of the BLI-4D isoform are identified within brackets and include a putative transmembrane domain, atrans-Golgi network localization signal (TGN) and a casein kinase II phosphorylation site (CSK II).











