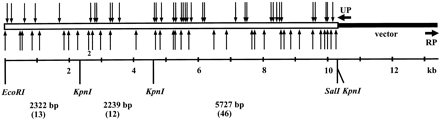
Distribution of cat–Mu integration sites in the plasmid pSR11. (Open bar) Mouse genomic insert; (arrows) transposon integration sites: Above the bar are indicated the transposons that contained the catgene in the same orientation as the Kcc2 gene; below, the transposons are in the opposite orientation. (RP, UP) Primers used to sequence the insert ends. The restriction sites used in cloning and screening of integrants are indicated. Also shown are lengths of the restriction fragments (in bp) produced by EcoRI–KpnI double digestion as well as the number of sequenced clones for each restriction fragment (in parenthesis).











