DNA Sequence Chromatogram Browsing Using JAVA and CORBA
- 1EMBL-Outstation—The European Bioinformatics Institute (EBI), Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SD, UK; 2Department of Computer and Information Science University of Pennsylvania, Philadelphia, Pennsylvania 19104 USA; 3Genome Sequencing Center, Washington University School of Medicine, St. Louis, Missouri 63108 USA
Abstract
DNA sequence chromatograms (traces) are the primary data source for all large-scale genomic and expressed sequence tags (ESTs) sequencing projects. Access to the sequencing trace assists many later analyses, for example contig assembly and polymorphism detection, but obtaining and using traces is problematic. Traces are not collected and published centrally, they are much larger than the base calls derived from them, and viewing them requires the interactivity of a local graphical client with local data. To provide efficient global access to DNA traces, we developed a client/server system based on flexible Java components integrated into other applications including an applet for use in a WWW browser and a stand-alone trace viewer. Client/server interaction is facilitated by CORBA middleware which provides a well-defined interface, a naming service, and location independence.
[The software is packaged as a Jar file available from the following URL: http://www.ebi.ac.uk/∼jparsons. Links to working examples of the trace viewers can be found athttp://corba.ebi.ac.uk/EST. All the Washington University mouse EST traces are available for browsing at the same URL.]
Biological Information Distribution
The Internet is host to an increasingly diverse range of mechanisms for biological data distribution. Two of the latest are the World Wide Web (WWW) standards set by the W3C (http://www.w3.org/), which are already well established among biologists, and the Object Management Group (OMG) Common Object Request Broker Architecture (CORBA), which is relatively new in this field (http://www.omg.org). The existing WWW standards have the advantage of simplicity and broad availability; however frequent extensions to HTML and additions such as JavaScript, XML, and dynamic HTML (See Box 1 for an aid to definitions. These are also all described athttp://www.hotwired.com/webmonkey/collections/crash_courses.html) have tested the WWW browser developers and the user’s ability to keep up. The incorporation of Java applets (http://java.sun.com) into HTML documents has further tested the maintenance of common standards as Java itself has undergone rapid change. However Java’s basic combination of security, portability, and desirability as a programming language have ensured the inclusion of a Java Virtual Machine (JVM) into the major WWW browsers where it increases the potential for client interactivity greatly. In 1996, the ease with which client/server object-oriented applications could be written, distributed, and supported across the Internet increased when Netscape (http://www.netscape.com/) announced (Orfali and Harkey 1997) that its browsers were all going to include a CORBA Object Request Broker (ORB), and when it decided subsequently to distribute its browsers for free.
The use of CORBA in a biological context was introduced by Hu et al. (1998) and Lijnzaad et al. (1998), who explained that CORBA can be a good solution to the problem of creating applications for distributed heterogeneous environments. The Internet is the extreme example of both distribution and heterogeneity and is described by Orfali and Harkey (1998) as being host to the Object Web in which CORBA and Java complement each other’s abilities to create globally accessible interactive objects. Principal among the benefits that CORBA brings to biological data distribution and interaction, are: the Interface Definition Language (IDL) to define interfaces between objects, scalability (including language and operating system independence), state-preservation across invocations, and a rich set of 15 object services, for example, the naming service.
Box 1.
Definitions
Sequence Chromatograms
DNA sequence chromatograms are interpreted to produce nucleotide sequences (base-calling) and corresponding base-call quality estimates but although these derived views are used more commonly, the traces remain the ultimate reference source for any queries about that particular sequencing reaction. All commonly used sequence assembly packages (for example Bonfield et al. 1995), include proprietary trace browsers to help the user (finisher) distinguish poor-quality data from good and so work backwards to recreate a representation of the original sequence. Furthermore, in regions with either trace artifacts specific to a particular sequencing chemistry, or general background contamination, an experienced finisher might be able to diagnose correctly the underlying problem and provide a better basecall when provided with a suitable view of the original trace. As with contig assembly, trace availability can increase the success rate of STS development from ESTs by enabling an optimal estimation of the possible positions of base-calling errors. Mott (1998) explored more of the direct uses of sequence traces through trace alignment including the identification of vector sequence (better than other automated methods), and detection and analysis of polymorphisms/mutations. Examples of existing trace viewers and editors include Ted (Gleeson and Hillier 1991), Consed (Gordon et al. 1998), and Trev (http://www.mrc-lmb.cam.ac.uk/pubseq/manual/trev_toc.html).
A poignant example in which the special role of sequencing traces as the ultimate sequencing reaction reference has emerged from the combination of the recent release of the Phred base-calling program (Ewing and Green 1998) and the status of the largest section of the public nucleotide databases: the EST sequences. ESTs are unusual in that they are submitted and published in a raw state with limited quality control (Hillier et al. 1996) and without the error detection and correction process intrinsic to normal shotgun assembly. Hillier et al. (1996) stressed the need for traces to be available online globally and have therefore maintained an ftp site where traces can be downloaded since the beginning of their EST sequencing. Now that Ewing and Green (1998) have released Phred with its improved base-calling (estimated to make 50% fewer errors than the original ABI base-caller) ∼250,000 entries in the GenBank (Benson et al. 1998) and EMBL (Stoesser et al. 1998) nucleotide databases may be considered to be out of date and ripe for replacement, whereas the original chromatograms remain available online and ready for reinterpretation at the originating laboratory.
Overall, the Internet is enabling decentralization within all areas of biological data access via the simplicity and low cost of HTML, the code portability of Java, and now the global middleware of CORBA. Though DNA sequence traces are collectively large, and scattered globally they are still important and following the same trends as other types of biological data: originally accessible via ftp, and now by Java applet over either HTML or the OMG’s Internet Inter-ORB Protocol (IIOP) as described in this paper.
RESULTS
A Java trace-viewing applet originally written by E. Buehler (see Fig. 1) has been developed into a set of trace-viewing tools with each component filling a different software niche. The tools work with different versions of the Java Virtual Machine; are packaged as Java applets, applications, and Java Beans, and operate as either CORBA client/server systems, or stand-alone applications.
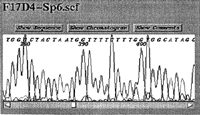
Screen shots of the original applet embedded in a web page (showing F17D4-Sp6), and the CORBA trace server client displaying a Washington University Genome Sequencing Center (WUGSC) EST trace (yx99h12.s1). Both clients offer scrolling, automatic scaling, and view selection of either the chromatogram, the called bases, or comments describing the conditions of the electrophoresis.
Design
The design choices were influenced by many factors including, most importantly, the fact that the majority of DNA sequence traces are normally stored in individual files in one of only three formats ABI, SCF V2, or SCF V3 (see Table 1). Most sequencing machines’ proprietary formats are convertible to the Standard Chromatogram File (SCF) formats (Dear and Staden 1992), a process helped by the Staden group’s provision of freely available SCF libraries (ftp://ftp.mrc-lmb.cam.ac.uk/pub/staden/src/). This lack of flat-file diversity enabled both the HTTP-based and CORBA-based trace-viewing clients to share much of the same code and also allowed a focus on scalability and download speed for the server design.
Summary of the Different Programs
The move from the original applet to the client/server design offers many benefits including: an object abstraction; the use of separately named trace stores, each with its own description; a choice of compressed, or uncompressed traces; and most importantly, the opportunity to generalize implementation details such as where and how a particular trace is stored to present a common database interface (see Box 2). The separation of client and server communicating through an agreed interface is the cornerstone of CORBA distributed software design allowing concurrent use of different languages and operating systems, yet allowing both clients and servers to improve implementations and add new features independently of each other. If, eventually, the original specification is found to be restrictive, new interfaces can be written and implemented, yet still supporting the old (unlike database schemas). The IDL language also allows inheritance so simple IDL specifications like that in Box 2 can be extended to create more complex derived interfaces. Downloading, parsing, and displaying a trace can take less than two seconds (from genome.wustl.edu in the USA to ebi.ac.uk in England) but may take more than five times longer when the Internet is congested (data not shown). Extra time is needed for an initial transfer of a Java ORB (if one is needed by the client). ABI format files are the largest and take the longest to arrive. SCF format trace files (either version) are already many times smaller than the original ABI format trace and SCF version 3 chromatograms can be compressed by gzip to <7% of the original file size. The SCF version 3 format was designed specifically to be compressed easily; seehttp://www.mrc-lmb.cam.ac.uk/pubseq/manual/formats_2.html.
Box 2.
The DNA Trace Database IDL Interface Specification
The software can be downloaded as either the compiled-class Jar files referenced from within the applet tags of any example applet pages (use view page source in Netscape), or as Java and IDL source Jar files as specified above. As an example not requiring Java or IDL compilation, nor a local ORB, one could use the local trace viewer to display the gzipped trace file mr32b07.r1.gz in the current directory with the command “java embl.ebi.trace.TraceView mr32b07.r1.gz” after the jar file containing all the chromatogram viewer classes is downloaded from an applet page and specified directly in the user’s local CLASSPATH environment variable. This jar file is typically called CorbaChromatogramApplet.jar and includes the TraceView.class file and all of its supporting classes. Thus, setting the environment for a UNIX csh session would require some version of a command such as “setenv CLASSPATH/home/myclasses/CorbaChromatogramApplet.jar.”
DISCUSSION
Currently, there a few problems in deploying CORBA-based applications over the Internet. These problems include: old firewalls blocking the IIOP protocol, the need to download ORB classes to clients because of the lack of a guaranteed local ORB, and a lack of support for multiple applet signing that would allow applets to follow object references to objects on computers other than the original applet’s host. There are already solutions to all these problems but their degree of irritation should decrease with the release of JDK1.2 from Javasoft (http://www.javasoft.com/) with its high-performance-class libraries and built-in Java ORB. When all operating systems support this rich environment, which includes OMG CORBA support, distributed computing may move further out of the browser and directly into more of a user’s normal molecular biology application set.
CORBA may appear to be overkill for this simple interface specification (relative to sockets for example) but as more biological software components are written to CORBA standards, any extra individual server installation effort becomes reduced. The EMBL outstation European Bioinformatics Institute (EBI) is working toward standards for such components along with other members of the OMG’s Life Science Research (LSR) Domain Task Force (DTF) (http://lsr.ebi.ac.uk/). Java RMI would have been an interesting CORBA alternative but was not investigated because of the lack of relevant biological standards efforts, frameworks, language independence, services, and local support.
Future Options
The trace viewer is limited by its isolation: Only when more CORBA servers are developed to support applications such as EST clustering, sequence assembly, etc., will the synergies of CORBA-wrapped data become obvious. The CORBA trace server will move to the new CORBA 3 standard, which supports fully portable (between different vendors’ ORBs) server code as soon as practical. The client should benefit from extra interactive features such as quality value display and editing, external trace view positioning interfaces, and multiple trace views.
METHODS
All the software is written in Java and compiled using Sun Microsystems/Javasoft’s Javac Java compilers (http://www.javasoft.com/). The simplest applet (also the first written applet) complies with the Java 1.0 standard but the remainder of the code requires Java 1.1 class libraries. The IDL interface specifications were compiled by Object Oriented Concepts’ (http://www.ooc.com/) ORBacus IDL to Java compiler. Many ORBs and IDL compilers are available free (http://industry.ebi.ac.uk/corba/) as are Sun’s Java compilers. Documentation is distributed throughout the code in Javadoc comments.
Implementation
The three trace formats are parsed by subclasses of an abstract chromatogram class. The chromatogram class visualization code is in a separate ChromatogramCanvas class to keep display and user-interactivity methods separate from the basic chromatogram object model. The client canvas uses double buffering to reduce flicker when scrolling. The chromatogram display can be switched to display ASCII base-calls, or the ABI sequencing machine’s comments field.
The client/server CORBA system wraps the client classes inside a CORBA adapter class. This adapter translates from GUI-generated trace load requests into CORBA method calls on a particular trace database server implementation via a CORBA naming service. The trace file parsing is done easily on the client because CPU cycles are plentiful, and the Java code transfer overhead is small (∼25% of the size of the smallest compressed trace). To optimize scalability and speed, the server can store and transfer traces as gzipped files which are handled easily by the Java.util.zip package in Java 1.1.
The CORBA trace server has all the implementation-specific methods for loading a trace (from a particular directory hierarchy or database) in a single class that can be overridden. The server configuration details including database names and descriptions are parsed from a simple text file that is read once when the server starts up. Multiple servers in different locations can register with a common naming service.
The original applet, being written to the older Java standard and using an ordinary http daemon as its download server is well suited to general Internet deployment in which browsers versions may be out of date and for small sequencing centers in which there are few traces for display and no local programming expertise. The implementation of Java 1.1 in Netscape Communicator 4.5 supports all the code described.
Acknowledgments
We are grateful to Tom Flores for helping to start the CORBA element of this project. Rodger Staden’s group, especially James Bonfield, helped with advice and support. This work was funded by European Union grant BIO 4 CT 960346.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵4 Corresponding author.
-
E-MAIL jparsons{at}ebi.ac.uk; FAX 44 1223 494468.
-
- Received October 6, 1998.
- Accepted January 20, 1999.
- Cold Spring Harbor Laboratory Press