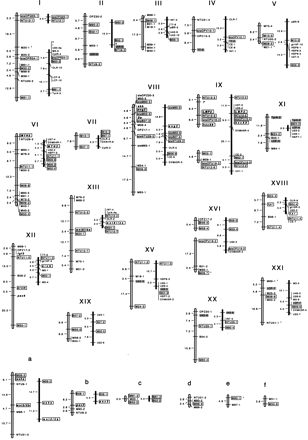
Genetic linkage map of medaka. The linkage groups on the left LGs I–XXI and LGs a–c are assigned by the female recombination-based mapping in this study. Those on the right are derived from Wada's male recombination-based map with anchoring markers. Numbers on the left of each linkage group indicate genetic distances (cM) calculated using the Kosambi mapping function. Marker names are shown at the right of each linkage group. Names of RAPD markers were given by the primer designation followed by dashes and the identification numbers of HNI-specific markers obtained with these primers. STSs were designated by the letters “sts” before the name of the original RAPD markers. The known genes and the Da locus are indicated in boldface type. The markers used for anchoring to Wada's map are boxed (white boxes are newly obtained markers in this study and shaded boxes are derived from Wada's markers). Numbering of the linkage groups was according to Wada et al. (1995). Six linkage groups (LGs a–f) could not be anchored in this study. Markers showing segregation distortion are indicated by asterisks.











