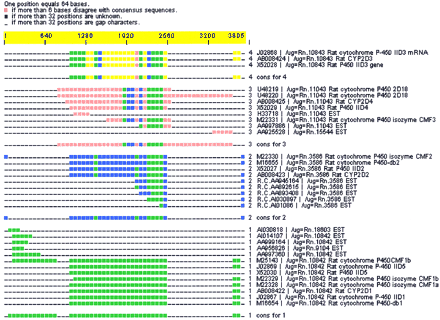
(A) CRAW report for a d2 cluster containing isozymes of mouse cytochrome P-450. Seven UniGene clusters (Rn.10843, Rn.3586, Rn.18603, Rn.10842, Rn.9104, Rn.11043, and Rn.15544) are merged. (B) (Online supplement available at www.genome.org andwww.pangeasystems.com) Interleaved multiple alignment showing a region of 240 bases with high identity alignment between all four cluster assemblies. d2_cluster has put all of these sequences together because of regions of high identity (as seen in Fig. 3B). UniGene has separated isozymes into distinct clusters, although UniGene clusters Rn.18603, Rn.10842, and Rn.9104 should probably form a single cluster according to reasonable clustering rules due to their perfect assembly into subgroup 1 and high overlap.











