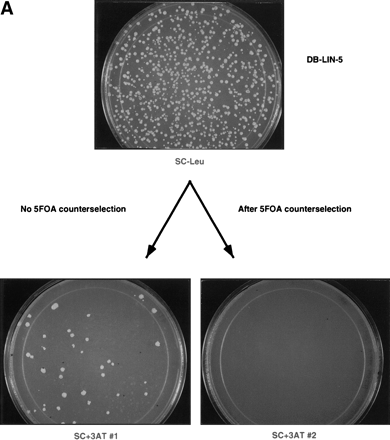
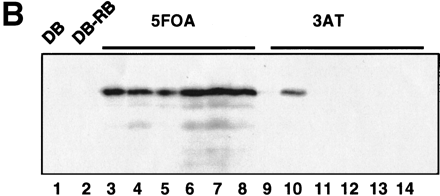
Preclearing of PCR-generated self-activator baits. (A) A PCR-generated lin-5 sequence was transformed by Gap repair into MaV103 cells. The Leu+ transformants were treated as described in the legend to Fig. 1. The plates shown here correspond to the Sc-Leu, Sc+3AT#1, and Sc+3AT#2 plates indicated in red in Fig. 1. (B) Western-blot analysis of extracts from yeast cells expressing self-activator (lanes 9–14) and non-self-activator (lanes 3–8) DB-LIN-5 baits. A mix of three anti-LIN-5 monoclonal antibodies that recognize epitopes in the LIN-5 carboxyl terminus were used. (Lanes 1, 2) Extracts from negative control cells expressing DB alone and DB-RB, respectively (RB corresponds to the pocket domain of the human retinoblastoma protein); (lanes 3, 9) extracts from pools of colonies; (lanes4–8, 10–14) extracts from individual colonies.











