
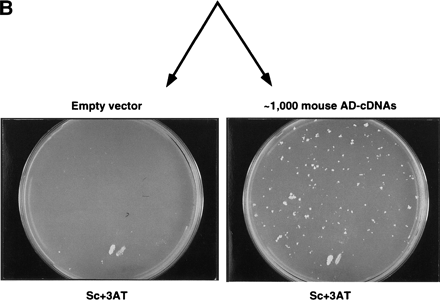
Preclearing of self-activator baits from random DB-cDNA libraries. (A) The mouse 13.5-day embryonic DB-cDNA library cloned in aLEU2 plasmid was transformed into MaV103 cells. The ∼50,000 resulting Leu+ transformants were treated as described in the legend to Fig. 1. The plates shown here correspond to the Sc-Leu, Sc+3AT#1, and Sc+3AT#2 plates indicated in red in Fig. 1. The Sc-3AT plate atright contains two control patches. (B) A mouse 13.5-day embryonic AD-cDNA library cloned in a TRP1 plasmid was transformed into MaV203 cells and the resulting transformants were mated with the precleared DB-cDNA transformants shown in A. The plate at right shows the resulting 3AT-growing colonies indicative of potential two-hybrid interactions. As a negative control, MaV203 was transformed with a TRP1 empty vector and mated with the precleared DB-cDNA transformants (left). Both plates contain two control patches at the bottom.











