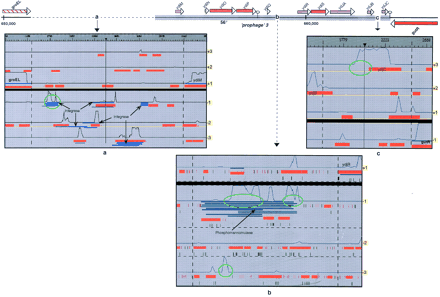
B. subtilis prophage 3 region containing three authentic frameshifts (a–c) corresponding to probable pseudogenes (see text). Boundaries of this prophage on the chromosome are indicated by the thick gray line at top. (a–c) Results of the analysis of these regions with the CoDing Sequences searching method (red boxes), the Blast2x method (blue rectangles), and the GeneMark coding predictions (black solid lines). (b) Start and stop codons are represented by pink and green lines, respectively. Atypical features are circled in green (respectively at positions 652000, 656000, 658500, 659000, and 663500 bp).











