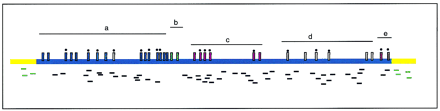
Schematic representation of the scanning procedure. Cosmid clone DNA (including vector sequence (yellow)) is sonicated, end repaired, and subcloned into EcoRV-cut pBluescript. Recombinant inserts are PCR amplified and sequenced from one end, generating ∼500 bp of sequence. These sequences are randomly distributed across the cosmid clone (small black bars). Low quality, vector, and E. colisequences are removed. One, 64-lane ABI377 sequence gel generates ∼50 good insert sequences providing 30%–50% coverage. An averageFugu cosmid clone will contain five to seven genes (represented as a–e); some are identifiable by BLAST homology at the protein level (fewer at the nucleotide level). The vertical colored bars represent the exons in the five genes and those with black dots above them are covered by the sequence scanning of this cosmid. Only the smallest genes are liable to be missed (geneb in this case). However, gene d has not been identified in other species as yet and so will not be recognized by BLAST searches (although gene prediction programs may do this) and some identified genes will show only low similarity with homologs across regions of the gene (e.g., the middle of gene a) and so may only give low BLAST scores.











