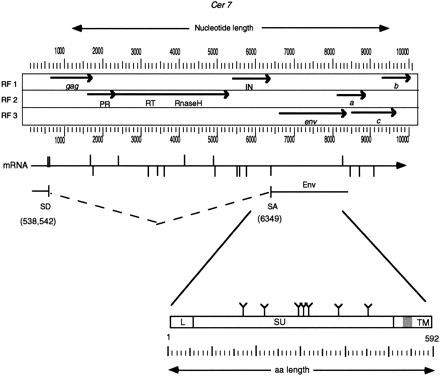
Cer7 ORF map and putative Env. The largest ORFs (>200 amino acids) from the three forward reading frames (RF1, RF2, and RF3) ofCer7 are translated and depicted as solid lines. Stop codons are shown as arrowheads. The common retroviral domains (PR and IN) and ORFS (gag and env) are labeled below the indicated ORFs. Note the three ORFs (a, b, and c) following the putativeenv ORF. Accessory proteins of complex retroviruses are found in analogous regions. The genomic mRNA is shown as a horizontal arrow below the ORF map. The predicted splice donor (SD) and splice acceptor (SA) sites are shown as vertical lines above and below the mRNA arrow, respectively. The predicted splice donor and acceptor sites that lead to the Env ORF production are shown below the mRNA. The Env protein is depicted as a box below the predicted spliced RNA. The leader peptide is indicated by an L. The possible N-glycosylation sites are depicted by a “Y” above the Env box. Vertical lines within the box represent putative protease cleavage sites that may serve to remove the leader peptide and cleave the region between the integral surface (SU) and transmembrane (TM) domains of Env. The predicted membrane anchor of TM is shown in grey. See Methods for a description of the functional motif predictions.











