
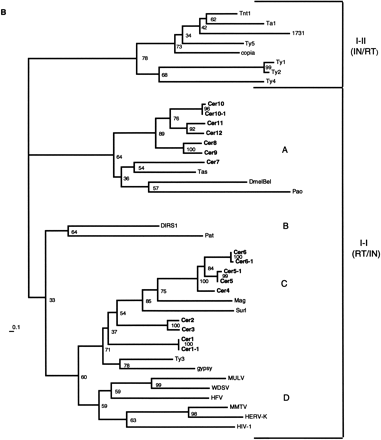
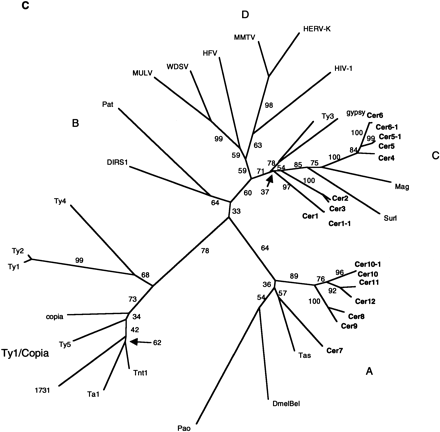
RT phylogenetic analysis. (A) RT amino acid alignment. The domains are boxed and numbered as by Xiong and Eickbush (1988). Additional RT sequences used in the Figure are referenced in Xiong and Eickbush (1990) or described as follows: (HFV) Human foamy virus, accession no. Y07725; (WDSV) walleye dermal sarcoma virus, accession no. AF033822; (PAT) Panagrellus redivivus retrotransposon, accession no. X60774; (Tas) A. lumbricoides retrotransposon, accession no. Z29712. Tas contains an in-frame termination codon at position 36 above. A gap was inserted at this position for the phylogenetic analysis. Residue coloring is based on the scheme described in Fig. 1. (B) NJ phylogram of sequences aligned inA (see methods). The tree was rooted with all of theCopia–Ty1 group LTR retrotransposons. Major clades are labeled according to the nomenclature proposed in Table 2. (C) Unrooted NJ phylogram of sequences aligned in A. Major clades are labeled as in B.











