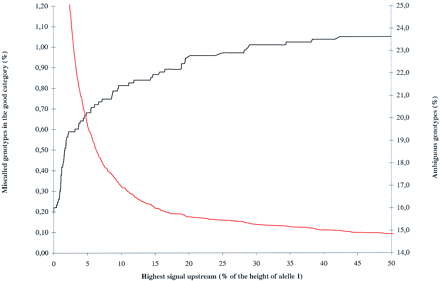
To capture genotypes that are erroneously called homozygous, but have an undetected peak upstream in the electropherogram, a function was incorporated in Decode-GT that detects homozygous allele calls, reads the intensity of the signal upstream from the called allele, and lists the genotype as ambiguous if the maximum value of the signal is above a defined value. The graph shows how the ambiguous category expands, and how the proportion of miscalled genotypes that are not included in the ambiguous category decreases as the threshold for the maximum height of the upstream signal is changed from zero to 50% of the height of the called allele. Based on this graph we decided to use 10% as the criteria for the highest signal. At that point, the ambiguous category is ∼17% and the miscalled genotypes are down to 0.8%. By going lower, the ambiguous category rises rapidly as does the number of captured miscalls.











