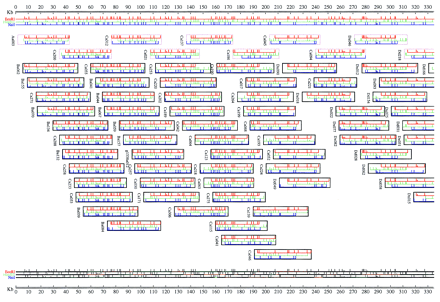
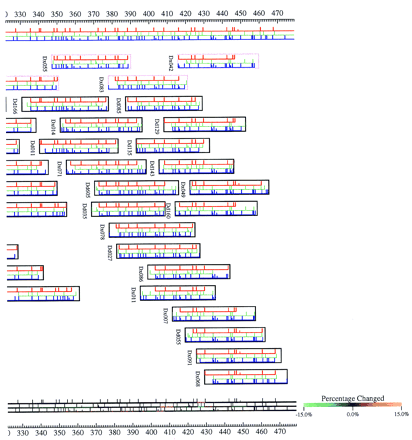
Depiction of one of several ways an individual user may view the end result of the ATLAS program and attempts to combine both the linear diagram for the MCD map being considered and the new difference system developed by ATLAS that is free of negative cycles. The restriction fragments are drawn for each enzyme map as horizontal lines bounded by restriction sites (vertical line segments). Group boundaries are indicated by taller vertical site marks, with the half-height site marks representing unordered fragments in the group. Inside the groups, the fragments are arbitrarily ordered by increasing fragment size. Clones are represented by rectangular boxes inside which the clone’s restriction fragments are drawn in alignment with the map. The clone boxes often extend past the ends of the first and last fragments in the clone reflecting total clone insert length that is factored into ATLAS’s clone end localization calculations. Because ATLAS modifies the bounds in the system of inequalities (which corresponds to changing both fragment and clone lengths), the final picture may contain fragments that have been stretched or shrunk in length from those in the actual MCD map. These changes often occur in regions of the map that contain mapping errors or may occur in regions in which many small target fragments have been missed. The duplicate MCD map drawn below the clones is color coded according to the percentage change in fragment length needed for ATLAS to draw the picture. Bright orange and green indicate that the fragments were either stretched or shrunk by 15% of their original length.











