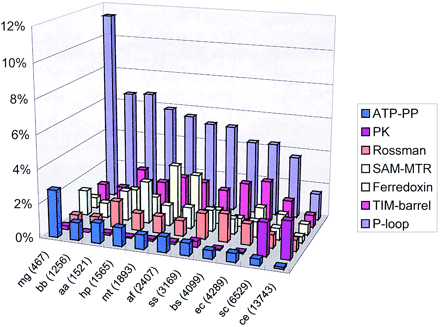
Distribution of the most common folds in selected bacterial, archaeal, and eukaryotic proteomes. (ATP-PP) ATP pyrophosphatases; (PK) serine–threonine protein kinases; (SAM-MTR) S-adenosyl methionine-dependent methyltransferases. (mg) Mycoplasma genitalium; (bb) Borrelia burgdorferi; (aa) Aquifex aeolicus; (hp) Helicobacter pylori; (mt)Methanobacterium thermoautotrophicum; (af) Archaeoglobus fulgidus; (ss) Synechocystis sp.; (bs) Bacillus subtilis; (ec) Escherichia coli; (sc) Saccharomyces cerevisiae; (ce) Caenorhabditis elegans. For each genome, the total number of encoded proteins is indicated in parenthesis.











