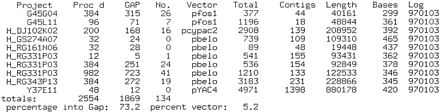
Typical daily sequencing report for a group. The report is indexed by sequencing project in the first column: The first two projects areCaenorhabditis briggsae fosmids, those prefixed by Hare human clones, and the remaining project is a C. elegansYAC. In order of appearance, the remaining columns show number of reads processed, number that entered the database, number which were vector sequence, name of the cloning vector, total number of reads currently in the database, total number of contigs currently in the database, basepair length of the clone based on the current assembly, average number of bases per read based on trace clipping, and the logging date. Below these are column totals: number of reads processed, number that entered databases, and number of vector reads. The last line gives overall percentages of reads that assembled and reads that were vector sequence. Information is reclassified for other reports, for example, weekly summaries are also indexed by the organism being sequenced. These reports assist group coordinators in tracking continuously the success of their sequencing group and in troubleshooting problems as they arise.











