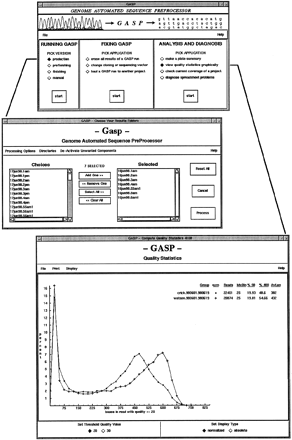
Graphical user interfaces. Main interface (top) serves as an integrated platform for all the preprocessing tools and is divided into three sections: preprocessor versions for specific types of reads (e.g., shotgun reads), error correction tools, and tools for analysis. As a result, users do not have to memorize numerous script names and command line options to use the toolkit effectively. Another interface (middle) is used to communicate input parameters to the sequence processing engine. The modified Unix environment is inherited from the main interface and two-way communication between the interface and the main program is handled via Perl’s IPC::Open2utility. A viewer for sequence quality statistics (bottom) enables inspection of results from a base quality standpoint. It has proven to be an effective means of monitoring the overall shotgun sequencing process and has come to play a role in evaluating changes in lab protocols.











