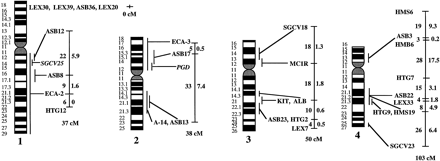
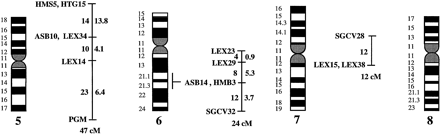
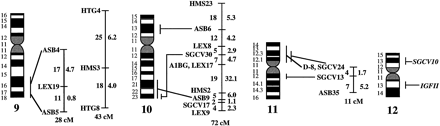
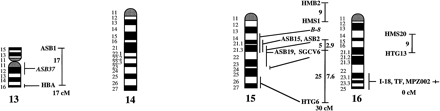
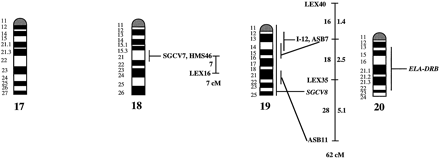
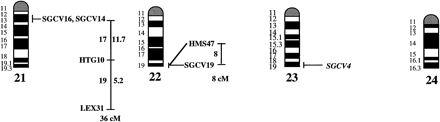
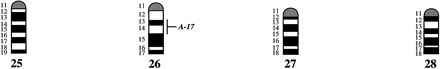
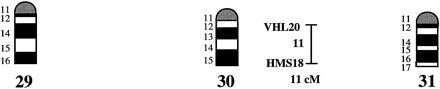
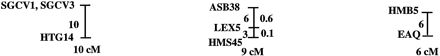
A preliminary male autosomal linkage map of the equine genome. The map depicts all established linkage groups anchored to chromosomes, as well as unassigned linkage groups (bottom). Chromosomal assignments of linkage groups are based on one or more of the markers being physically mapped by in situ hybridization (vertical bars close to chromosomes). The only exceptions are the LEX30–LEX39–MPZ027–LEX20 linkage group on ECA1, the HMS5–HTG15–ASB10–LEX34–LEX14–PGM linkage group on ECA5, the SGCV28–LEX15–LEX38 linkage group on ECA7, the HTG4–HMS3–HTG8 linkage group on ECA9, and HMS20–HTG13 on ECA16, which chromosomal assignments are based on synteny data (Shiue et al. 1998; see also Bailey et al. 1997; Godard et al. 1997). The assignment of the HMB2-HMS1 group to ECA15 is based on a previously observed linkage between one of these markers and physically anchored markers on this chromosome (Godard et al. 1997). The assignment of the group on ECA30 is based on a defined aneuploidy (Bowling et al. 1997b). Values to the left of main vertical bars represent genetic distances between markers expressed as multipoint Kosambi cM, and values to the right are the log 10 odds against reversed order of adjacent markers. Loci shown at the same vertical position had 0% recombination. Below each linkage group is indicated its total multipoint length. In situ mapped markers genotyped in the family material but not showing linkage are depicted in italics on the map according to their band assignments; their exclusion from linkage groups is evident from the absence of a horizontal line in the vertical bar connecting linked markers. Three markers (ASB13 on ECA2, MC1R on ECA3, and ASB11 on ECA19) known from in situ hybridization to reside within established linkage groups were included in multipoint analyses although only showing a maximum two-point lod score of between 2 and 3.











