WebWise: Guide to the Joint Genome Institute Web Site
This installment of the WebWise series reviews the Joint Genome Institute (JGI) web site (http://www.jgi.doe.gov/). The JGI, established in 1997 (Casey 1996), represents an ongoing consolidation of the U.S. Department of Energy (DOE) genome sequencing centers established at the Los Alamos (LANL), Lawrence Berkeley (LBNL), and Lawrence Livermore National Laboratories (LLNL). The JGI web site presents a centralized information center to disseminate information pertaining to the sequencing efforts still being carried out at the three laboratories while a new central research facility is being built. Many of the data links lead to pages that are actually still hosted by these three research laboratories.
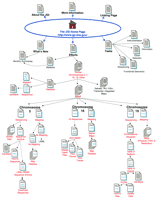
This review is intended to summarize the consolidated JGI web resource; pages hosted by LANL, LBNL, or LLNL are indicated on the site map displayed in Figure 1. However, the site map does not attempt to represent all of the information available at the LANL, LBNL, or LLNL web sites (http://www-ls.lanl.gov/;http://www-hgc.lbl.gov/;http://www-bio.llnl.gov/bbrp/genome/genome.html). The site map (Fig. 1) is intended to provide a simple road map to data and tools pages. Therefore, it does not include all of the links provided on the JGI site map; redundant links as well as some groupings of related links are depicted as a single link on the site map. The main features of this web site, as well as the features of those sites already reviewed, are indicated in Table 1. As with the previous WebWise reviews, you may find it useful to point your Browser window to the JGI web site while reading.
Site map for the JGI. The main links to pages discussed in the text are illustrated here. Links placed above the Home Page icon are to general informational resources; the links located on the bottom portion are to the data and software tools. Some links available on the web site are not indicated here.
Features of One JGI Web Site
General Information
The JGI Home page (http://www.jgi.doe.gov/) presents a simple, effective organization. Although there are numerous pages associated with this web site, they are grouped into six general publicly accessible sections (a seventh area is for in-house use only). One of these sections, namely the Linking Page area, still needs to be developed. In this review the term “section” refers to the text displayed on the left side of the web page, and “subsection” indicates text located to the right of each section title. Each section represents a branch point from the Home page. Many of the primary links or subsections available from each section page are also listed on the Home page. This design style facilitates navigation, as it is easy to determine the type of information available via each general section. In addition, this design allows one to jump directly to a more internal page, which is a useful feature for those aiming for a specific inner region of the web site.
All of the pages that are hosted by the JGI web server present a uniform design style. These pages group each subsequent layer of links into general sections. Each section, highlighted in alternating background colors of gray or white, includes links to the next set of pages that relate to that section title. A set of navigation links to each of the main sections included on the Home page, and an e-mail link to the JGI web master, are included on the bottom of each of these pages. Some inner pages include links leading to pages hosted by the three national laboratories; these pages present different design styles and do not consistently include links back to the JGI web site. It is a large undertaking to integrate access to data generated at LANL, LBNL, and LLNL; the first few layers of the JGI web site succeed at pulling the three laboratories into a single web focal point. Obviously some remaining integration still needs to be completed by adding links from the national laboratory web pages back to the JGI web site.
Two of the Home page section links lead to some general information about the JGI. Follow the About the JGI link to access information about organizational structure, purpose, publications, and staff (http://www.jgi.doe.gov/JGI_about.html). The links grouped under the Purpose and Organization sections of this page lead to some general background information about the JGI and the Human Genome Project (HGP). More detailed information about the latter is provided via the last two links in the Purpose section (Human Genome Project and History of DOE HGP). An organizational diagram, as well as lists of management and other staff members, is available in the People section of this page. These personnel pages include phone numbers and e-mail links. The Current Documents section includes links to documents defining the JGI purpose, sequencing effort, quality standards, and the production sequencing facility. The Production Sequencing Facility page includes an overview of the relationship between this facility and the national laboratories, as well as facility specifications.
The More Information Home page link leads to a page that includes General Information, For Students, For the Media, For Businesses, and Job Opportunities sections (http://www.jgi.doe.gov/JGI_info.html). The General Information section includes some links also listed on the About the JGI page but also presents links to a Molecular Genetics primer, information on Ethical Issues, and a FAQ page. Links in the media and business sections of this page lead to empty pages still under construction at the time of this writing.
Data
All of the JGI map and sequence data pages can be reached via the Efforts section link on the Home page. The page, entitled Chromosome Efforts for the Joint Genome Institute (http://www.jgi.doe.gov/JGI_efforts.html), includes links to sections on Statistics, Chromosomes 5, 7, 16, 19, and Other Efforts. The links in the Statistics section are apparently intended to provide a general overview of the whole JGI effort. Unfortunately, the Regions Mapped and Regions Sequenced pages are not yet available; hopefully these pages will provide an overview of the chromosomal regions being sequenced as well as the amount of data accumulated per chromosome. The Monthly Summaries page (http://www.jgi.doe.gov/Docs/JGI_Seq_Summary.html) presents a table indicating the amount of sequence accumulated over time and as compared to the goals set. These data are further broken down to reflect the difference between goals and rate of achievement for data submission versus full closure. This page can also be reached from the What’s New section of the Home page.
The Human Chromosome 7 and Other Efforts sections reflect the fact that the JGI web site is still under construction. Most of the pages called up by these section links display a warning that the page is “Not Ready Yet!”. The one link that does not point to this page, namely the P1 sequencing link in the Other Efforts section, does not lead to the page intended. This link should be updated to point to the correct page on the LBNL rearranged web site.
Following the Chromosome 5, 16, or 19 links brings up individual chromosome section pages that are consistently organized. For the most part the chromosome subsections provided include Efforts, Statistics, and several genetic and physical mapping sections. These mapping sections include many links to different sources for Genetic maps, Radiation Hybrid (RH) maps, FISH maps, Transcript maps, External Sequencing resources, and Integrated maps. Obviously the exact nature of the links provided on the Chromosome 5, 16, and 19 pages reflects the amount of additional data available for these three chromosomes. The External Sequencing Resources section is somewhat limited; the two links provided lead to a Human Genome Organization (HUGO) overview of sequencing activities per chromosome and to a mysterious “forbidden access” page at The Institute for Genomic Research (TIGR). While acknowledging that the map sections provide a very useful collection of chromosome resource links, because these links are not directly related to the sequencing effort under way at the JGI, they are not described in depth here.
The Statistics and Efforts sections provide indications of the amount of mapping and sequencing data completed and still “in the pipeline”, respectively. The links provided in these sections lead, for the most part, to web pages hosted by the LANL, LBNL, and LLNL rather than the JGI. The Statistics subsection Mapped links all lead to the Not Ready Yet! page. One nice feature of the pages reached via the Sequenced link (follow the links from the Statistics section of the pages devoted to chromosomes 5, 16, or 19) is the consistent display presented by the three laboratories. The sources and current locations of these pages are indicated in Table 2. The three laboratories display the sequenced, or completed project, data in a tabular format that includes clone name, contig name, accession number (linked to the GenBank record), number of bases, number of unique bases, and phrap values. Projects completed are tallied on a monthly basis, and separate tables are provided for each month. One useful addition to the LLNL table is the inclusion of cytogenetic band information. This provides at least a limited indication of how a given contig relates to the map data. Altogether, these pages could benefit by being kept more consistently up to date. For instance, the most recent month included on the LBNL Chromosome 5 page, at the time of this writing, was for February 1998. Although these tables do exhibit some minor style differences in color scheme and column order, the generally consistent style makes it convenient to work with these data.
The Location and Source of Chromosome-Specific Sequence Data
Unfortunately this effort to adopt a fairly uniform display format does not extend to pages concerning data still in the pipeline (reached via the Efforts section of each chromosome-specific page). The map and sequence displays available for the chromosome 5, 16, and 19 efforts are quite dissimilar in terms of accessibility, user interface, and display. The locations and laboratory sources of the JGI mapping and sequencing data for chromosomes 5, 16, and 19 are indicated in Table 1.
Chromosome 5
The chromosome 5 map displays, hosted by the LBNL server, are organized into sections. Even so, these maps are excessively large and difficult to view; both vertical and horizontal scrolling must be utilized to view the whole map. The display is not image-mapped, and clones used for sequencing are colored in red and associated with a name prefixed by “H.” Unfortunately, the map display style utilized impedes efforts to determine the correspondence between map and sequence data.
Follow the Sequencing link in the Efforts section of the Chromosome 5 page to access the sequence data. The page called up includes several links to pages presenting data quality information; the Sequencing Archive link leads to tables of clones sequenced, or in progress, at the LBNL. This page does not present a completely up-to-date view of the sequencing in progress as it is not updated on an automatic daily basis. A table of sequences submitted to GenBank is presented at the top of this page and below that there is a large table of clones currently in the pipeline. These two tables include the clone names, links to the GenBank record, links to LBNL local FASTA files of the completed or contig sequences, and an indication of the sequence size. If you follow the link to the LBNL/BDGP Sequence Archive and select the Human link on the following page, you will be presented with the option to download all of the human contig sequence data as a compressed multiple FASTA file.
Chromosome 16
The chromosome 16 map displays are located at the LANL; the URL of the top-level LANL map page is listed in Table 2. These web pages present multiple options including queries of the chromosome 16 database, a feature that was not tested for this review. From the JGI Chromosome 16 page, follow the Mapping link, then the Sigma Maps link to reach a page presenting links to several different maps (http://www-ls.lanl.gov/map_image.html). The P and Q arm contig maps are more condensed than the maps seen for chromosome 5. One would expect the P-Arm and Q-Arm Contig Maps to include some indication of the sequencing clone tiling path. Unfortunately, these maps lack sufficient information to support an “at a glance” determination of which clones are used for the sequencing effort.
The chromosome 16 sequence data are reached by following the 16 Sequences link from the JGI Chromosome 16 page (Efforts section). The page reached (the URL is indicated in Table 1) loads a Java Applet, so be sure that you have enabled this feature in your web browser. The initial Applet presents a series of buttons whereby you can opt to View Clones, View Traces, Display FASTA Files, SCAN Results, or follow links to other pages. Space considerations forestall providing an in-depth description of all aspects of this Applet here. However, suffice it to say that the Display FASTA Files option does result in displaying the FASTA sequences obtained for a selected clone in your Browser window. Of course, it is not an easy matter to decide which clone you might be interested in, as the map and sequence data are not closely integrated on these pages. The mere fact that a View Clones button is provided contributes to the initial hope that this might lead to a clone tiling path display. Disappointingly, this is not the case; furthermore, this part of the Java Applet did not work in an intuitive manner—repeated attempts to select the right combination of library, clone name, and display option only resulted in opening up numerous Applet windows that were then difficult to close. Items must be selected in the correct order: One must select a library, then a clone, and wait for a minute or two before proceeding. An assembly file is loaded during the waiting period and data in the file are then used to output one of the display options. The assembly file is not provided for many of the clones, in which case a window appears containing an error message indicating that the file could not be opened. To see a working example, select library hs16c and clone RT247.
Chromosome 19
The chromosome 19 map and sequence data are generated at the LLNL (see Table 2 for the LLNL Mapping page URL). Follow the Mapping link from the Efforts section on the Chromosome 19 page to access physical, FISH, and restriction map data. The LLNL Mapping page presents some general background information on the top of the page; scroll down to reach the links to specific maps. The physical map displays are presented in a vertical orientation and so conveniently do not require the use of both vertical and horizontal scrolling to view. Maps include an indication of the regions where contigs are sequenced or in the sequencing queue (the thick, vertical blue bars); unfortunately, it is not immediately obvious to the casual or first time visitor what the clone names are. The legend indicates the cosmid clone names are listed vertically but does not explicitly indicate what the other nonsequential numerical labels mean, which hampers interpretation.
The chromosome 19 map data can be reached by following the Sequencing link, from the Efforts section, on JGI’s Chromosome 19 page, which leads to a page display of the chromosome 19 ideogram (see Table 1 for the URL). The resulting graphic is image-mapped, and one must simply select a band of interest to click on. This action leads one to a Table of (Sequenced) Regions, which in turn is linked to a page that presents a Table of Clones for which sequence data are available. These tables include links to the GenBank report, and the clone name calls up a page presenting Contigs, in tabular format, from which you can then access either the LLNL FASTA file or Quality Data about that contig. This approach calls for several navigational jumps from page to page but otherwise seems a reasonable approach to displaying these data.
Tools
The JGI web pages offer very little in terms of software tools at this time. They do provide some descriptions of informatics and instrumentation goals (follow the Tasks link from the Home page (http://www.jgi.doe.gov/JGI_tasks.html#_Informatics). The Informatics section of this page provides links to general overviews of the research tasks being supported by computer software tools that are either currently in use or in development. The Research (OHER) link in this section brings you to a page containing links to additional informatics research information sources. Altogether, a lot of information is provided here. However, there are no in-depth descriptions of software tools and no links are provided to test, download, or use software tools of any type.
Although a Search link is provided at the bottom of each page, these links merely bring you to a page indicating the tool is not available yet. On the plus side, the presence of this link does indicate that searches will be supported at some point in the future. This is an important service to provide for a web site of this size and hopefully it will become available in the near future. The JGI web site also fails to offer a BLAST server at this time. Although the LANL, LBNL, and LLNL web sites were not examined in minute detail, there is no obvious indication that any of these sites currently provide, or plan to provide, this service. The current task to combine data and resources from these three centers is a large undertaking—disseminating these data by means of a BLAST service would be a convenient and flexible way to provide a more integrated public view of the JGI sequence data.
Conclusions
The JGI web site is a fairly recent addition to the host of HGP-related web sites. As such, they have taken advantage of the currently recommended approaches to designing informational web sites. The first several layers of the web site are well organized, provide consistent navigational links, and adopt a uniform design style. On the other hand, the inner data pages do not present a consistent organization or style and for the most part lack navigational links to the JGI or other pages. One hopes that this is merely a reflection of the current status of the integration task and that with time the data pages will also become more consistent and navigable. Furthering the integration of the LANL, LBNL, and LLNL into the JGI in this manner will increase the overall utility of the JGI web site and the data itself.
Although a large amount of map and sequence data can be reached via the JGI web site, these data are not tightly linked and so it is not easy to discern the relative order of sequenced clones or the general region to which these clones correspond. The image-mapped chromosome 19 ideogram does at least provide a general indication of the region to which a set of clones corresponds, but it is more time consuming than it needs be to determine the clone-to-map correspondence for the chromosome 5 and 16 sequence data. The utility of the sequence data is further hampered by the reduced frequency updates of some of the data web pages. Text at the top of the Chromosome 19 Sequencing page indicates these pages are a current reflection of the data accumulated, but the Chromosome 5 Sequencing page is updated only once a week. An explicit indication of the update frequency for the Chromosome 16 Sequence data pages is not provided.
Clearly, this web site is still under development, yet the utility of offering an integrated view of the DOE sequencing effort is apparent. Although there are some areas of the web site explicitly identified as incomplete, it is not clear to what extent the sequence data displays will be further integrated and modified. Public access to, and the overall utility of, these data will be enhanced if the integration effort includes the data display pages and if additional data interface resources, such as image-mapped contig maps and BLAST analysis, are provided in the future.
Footnotes
-
↵1 Corresponding author.
-
Next Month: The Institute for Genome Research
-
E-MAIL pruitt{at}ncbi.nlm.nih.gov; FAX (301) 435-2433.
- Cold Spring Harbor Laboratory Press