Direct Selection of Conserved cDNAs from the DiGeorge Critical Region: Isolation of a Novel CDC45-Like Gene
Abstract
We have used a modified direct selection technique to detect transcripts that are both evolutionary conserved and developmentally expressed. The enrichment for homologous mouse cDNAs by use of human genomic DNA as template is shown to be an efficient and rapid approach for generating transcript maps. Deletions of human 22q11 are associated with several clinical syndromes, with overlapping phenotypes, for example, velocardiofacial syndrome (VCFS) and DiGeorge syndrome (DGS). A large number of transcriptional units exist within the defined critical region, many of which have been identified previously by direct selection. However, no single obvious candidate gene for the VCFS/DGS phenotype has yet been found. Our technique has been applied to the DiGeorge critical region and has resulted in the isolation of a novel candidate gene, Cdc45l2, similar to yeast Cdc45p.
[The sequence data described in this paper have been submitted to the EMBL data library under accession nos. AJ0223728 and AF0223729.]
Deletions of chromosome 22q11 are among the most common structural chromosome abnormalities in man, with an estimated incidence of 1 in 4000–5000 newborns (Scambler 1993). The phenotypic spectrum associated with these deletions is broad and includes DiGeorge syndrome (DGS) and velocardiofacial syndromes (VCFS) (de la Chapelle et al. 1981; Scambler et al. 1992; Budarf and Emanuel 1997). Individual deletions are usually large, encompassing a commonly deleted region of 2–3 Mb (Lindsay et al. 1995; Carlson et al. 1997). Mapping of balanced and unbalanced translocation breakpoints has identified the proximal part of the deletion as the DiGeorge critical region (DGCR) (Augusseau et al. 1986; Li et al. 1994; Budarf et al. 1995; Demczuk et al. 1995;Levy et al. 1995).
We and others have established genomic contigs spanning the DGCR (Gong et al. 1996; Carlson et al. 1997) and a number of transcripts have been identified from this region (Budarf and Emanuel 1997). To date, none of the transcripts mapped to the DGCR have been shown to be mutated in nondeleted VCFS/DGS patients and it remains unclear whether the phenotype is the result of haploinsufficiency of one or multiple genes. It is therefore important to identify all transcripts from this region that may have a developmental role, and to this end, we have modified the direct selection technique to detect additional transcripts that are evolutionary conserved and developmentally expressed. Direct selection of conserved genomic DNA sequences has been described previously (Sedlacek et al. 1993), but this is the first report of conserved cDNA selection. Using this technique we were able to identify a novel gene that is similar to the yeast cell division control protein, Cdc45p (Hopwood and Dalton 1996). The human homolog of this gene maps to the distal part of the DGCR.
Comparative mapping between human 22q11 and MMU16 is well delineated (Botta et al. 1997; Galili et al. 1997), and, in another model organism, the Japanese pufferfish, Fugu rubripes (Brenner et al. 1993), genomic clones containing DGCR transcripts such asIDD (Demczuk et al. 1995; Wadey et al. 1995), HIRA(Halford et al. 1993), and ES2 (Rizzu et al. 1996) have been isolated (Taylor 1997). In a further extension of the direct selection technique, we used these Fugu rubripes genomic clones to select highly conserved and developmentally expressed murine cDNAs. Murine homologs of IDD and HIRA were selected, showing the power of conservation cDNA selection.
RESULTS
Isolation and Analysis of cDNA Clones: Human vs. Human
The cDNA selection experiments were divided into three groups (Fig. 1), covering a total genomic region of ∼450 kb and including the entire DGCR with the exception of a region spanned by HIRA. Initially, conventional direct selection was performed (see Methods) over ∼250 kb, spanning from IDD toHIRA by use of human fosmids and random primed human fetal brain cDNA (group 1A). In total, 223 cDNA-selected subclones were analyzed either by hybridization with positive controls or by sequencing and database searches (Table 1; column 2). The proportion of background sequences was low (3.4%), with the majority of clones being known genes. All five known positive control genes, IDD, ES2, CTP (Goldmuntz et al. 1996), HIRA,and CLTD (Sirotkin et al. 1996) were present in the cDNA select subclones as determined by colony hybridization and by specific EST sequence (data not shown). Two novel serine/threonine kinase genes,TSK and TSKP were also detected: Subsequently,TSK was also cloned by cDNA selection (Gong et al. 1996) andTSKP was predicted by comparative sequence analysis of human 22q11 and MMU16 (Goldmuntz et al. 1997). The complete genomic sequence from the DGCR, deposited into the GenBank database (M. Budarf, F. Chen, B. Emanuel, A. Hua, N. Ma, B.A. Roe, S. Toth, Z. Wang, G. Zhang, unpubl.), has permitted accurate mapping of the cDNA select subclones (ESTs) by sequence comparison: Approximately 16% of the clones (lone ESTs) mapped to the selected region but are unlikely to represent genuine ESTs, being single nonoverlapping, unspliced transcripts with limited ORFs. Lone ESTs have been reported in other direct selection experiments (Hattier et al. 1995; Ruddy et al. 1997) and are likely to arise from heterogeneous nuclear RNA transcripts or contaminating genomic DNA.
Schematic of DGS critical region on human chromosome 22q11 covering ∼500 kb. (Broken vertical lines) Patient translocation breakpoints. Transcripts are in italics, and their transcriptional orientation is shown by arrows at bottom. (Solid lines) Regions covered by the direct selection.
Summary of ESTs Generated from Direct Selection
Isolation and Analysis of cDNA Clones: Human vs. Mouse
Conservation cDNA selection is a modification of the direct selection approach with two main differences in methodology: First, DNA from two different species is used; and second, the captured biotinylated genomic DNA/cDNA hybrid is washed at a lower stringency prior to cDNA elution. For comparison with the initial human versus human-direct selection, a similar area was covered by conservation cDNA selection (see Methods) by use of human fosmids and random primed 13.5 and 14.5 days postcoitum (dpc) mouse fetal cDNAs (group 1B). The low stringency (LS) products were subcloned and analyzed (Table 1, column 3), and the high stringency (HS) products were radiolabeled and hybridized to PCR panels of LS subclones to identify strongly conserved ESTs (see Materials and Methods; data not shown). The relative background is very low, with the vast majority of clones (87%) representing known genes. All positive controls were present apart from the CLTD gene. No novel genes were detected as the selected genomic area excluded TSK and TSKP. However, 7% of the ESTs represented transcripts for two genes, the ribosomal L34 gene (human L34 gene GenBank accession no. L38941) and the keratin D gene (Rommens et al. 1995) and sequence comparisons with the selected genomic region revealed the presence of two potential corresponding pseudogenes (data not shown). In contrast to the same species selection result, no lone ESTs were subcloned.
A second conservation cDNA selection experiment (group 2), encompassing a region from HIRA to the 5878 breakpoint (∼200 kb), was performed. The results (Table 1, column 4) showed a much higher background level as a result of Escherchia coli DNA contamination during subcloning, but the ESTs generated gave a similar pattern as for group 1B: All three positive controls, Ufd1l(Pizzuti et al. 1997), Tmvcf (Sirotkin et al. 1997), andHira, were present and no lone ESTs were selected. Twenty-four percent of the ESTs represented a cDNA contig encoding a novel gene designated Cdc45l2.
Isolation and Characterization of Cdc45l2
A Cdc45l2 EST was used to isolate a full-length mouse cDNA clone (EMBL accession no. AJ223729). The sequence contained an ATG start codon within a region that conforms to the Kozak consensus sequence (Kozak 1996), followed by an ORF of 1704 bp. Two human ESTs, identified by database screening with Cdc45l2 (see Methods), were sequenced and found to be highly homologous to Cdc45l2,and represented full-length human CDC45L2 transcripts (EMBL accession no. AJ223728). CDC45L2 has an ORF of 1701 bp and shows 92.2% similarity (89.2% identity) to Cdc45l2 at the amino acid level. Database analysis revealed that the CDC45L2transcript consisted of 19 exons spread over a region of 40,662 bp between the UFD1L and TMVCF genes and is transcribed in a centromeric-to-telomeric direction.
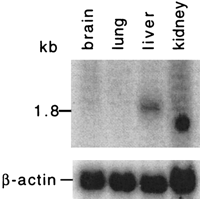
Northern analysis revealed a single 1.8-kb band in human fetal liver only (Fig. 2). CDC45L2 ESTs have also been found in libraries derived from an 8-week human embryo, fetal liver and spleen, and T-cell, testis, and pancreatic tumor. Cdc45l2cDNAs have been isolated from 3.5–14.5 dpc embryonic murine and macrophage libraries.
Northern analysis of Cdc45l2. A single 1.8-kb band was seen in the human fetal liver lane. β-Actin is shown as a loading control.
Database screening revealed similarity to just three other proteins: yeast Cdc45p (Hopwood and Dalton 1996; Hardy 1997; Zou et al. 1997); Tsd2 protein from the smut fungus, Ustilago maydis (Onel and Holloman 1997); and a Caenorhabditis elegans hypothetical protein F34D10.2 (TREMBL accession no. Q19998), with probabilities of 1.4 e −30 to 3.4 e −40. Figure 3 shows a Clustal W (Thompson et al. 1994) alignment of the four proteins.
Conservation cDNA Selection Between F. rubripes andMus musculus
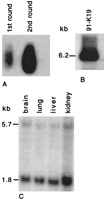
To assess the limits of conservation cDNA selection, a pilot study was performed by use of the more evolutionary distant model organism, the Japanese pufferfish, F. rubripes. An F. rubripescosmid, 91-K19, containing a homology of synteny grouping with the DGCR was identified by hybridization with IDD and ES2(Taylor 1997). Conservation cDNA selection was conducted under the same conditions as above, and hybridization analysis revealed the presence of Idd but not Es2 in the cDNA selection PCR products (Fig. 4A). Comparison of the F. rubripesgenomic DNA sequence with the murine cDNAs for Idd andEs2 revealed a much higher homology for Idd than forEs2 (data not shown), which could account for the nonselection of the latter. The second round low stringency PCR products were subcloned and sequenced (Table 1, column 5): The background consists mainly of globin cDNAs and the majority of the ESTs are present in dbEST. The known genes include not only Idd but also others identified by direct sequencing of 91-K19, for example, histone H3 and a subunit of cytochrome c oxidase (Taylor 1997). One novel gene, Fug1, identified by 5% of ESTs, has been mapped back to 91-K19, and gives two transcripts on a human fetal Northern blot (Fig.4B,C). A similar experiment by use of an F. rubripes cosmid identified by cross-hybridization to Hira, selected murineHira cDNAs as well as a number of other unknown ESTs.
Results from F. rubripes vs. mouse cDNA selection. (A) Southern blot of first and second round cDNA selection PCR products hybridized with radiolabeled Idd, washed to a stringency of 0.1× SSC, 0.1% SDS at 65°C. (B) Southern blot of BamHI- and EcoRI-digested cosmid 91-K19 hybridized with radiolabeled Fug1 EST. (C) Northern analysis with a Fug1 EST. Two bands (5.7 kb and 1.8 kb) are seen in all lanes. β-Actin, as a loading control, is shown in Fig.2.
DISCUSSION
Our results from the direct selection of the DGCR illustrate the power of this technique with same species selection (group 1A) generating ESTs for all known genes in the area except GSCL(Gottlieb et al. 1997). Conservation cDNA selection (group 1B), covering a similar genomic region to group 1A, also generated ESTs for all the known genes except CLTD, which is unlikely to be conserved (Gong et al. 1996; Botta et al. 1997), and Gscl(Galili et al. 1997). However, a retrospective analysis of the total group 1 PCR products, by Southern hybridization with a humanGSCL genomic fragment, revealed the presence of Gsclin the group 1B selection products: PCR analysis with specific primers for Gscl detected the presence of a full-length mouse transcript in the low stringency cDNA products from group 1B (G. Lee, unpubl.). Failure to detect GSCL in the human group 1A PCR products may be accounted for by differences in cDNA source and may show one of the advantages of conservation selection: The type of cDNA source allows access to developmental stages and tissues for genes expressed in a narrow developmental window, even those likeGscl, expressed at very low levels.
Conservation cDNA selection can be considered a very useful adaptation of the direct selection approach for a number of reasons in addition to the above: (1) The selective cloning of conserved coding regions allows a more rapid identification of gene families/motifs; (2) the ESTs obtained are immediately suitable for embryo expression analysis; (3) lone ESTs should not be selected; and (4) the ESTs are useful for comparative mapping purposes. For example, because coding regions are preferentially cloned, such sequences are useful in the derivation of CATS (Lyons et al. 1997). Some potential drawbacks of the system are (1) transcripts specifically expressed in humans are missed; (2) the lack of ESTs encoding the less well-conserved 3′ untranslated region (UTR), and a concomitantly decreased chance of matching sequence in the databases (such as dbEST) that is often 3′ UTR; and (3) very strongly conserved regions of gene families that map elsewhere could theoretically be selected.
The preliminary findings from conservation cDNA selection betweenF. rubripes and mouse highlights both the power of the technique and its limitations. Two of three known genes in the region were cloned, with the most weakly conserved gene being missed. However, there was a very high background of globin ESTs and evidence that strongly conserved motifs of gene families (e.g., cytochrome coxidase) were being selected. To overcome these limitations, several steps could be taken such as (1) incorporating further rounds of selection (Sedlacek et al. 1993); (2) varying the washing stringency of the genomic DNA/cDNA hybrid molecule; and (3) blocking the background of globin ESTs by hybrization of cosmid genomic DNA with excess globin cDNA in addition to Cot1 DNA.
Conservation cDNA selection products from group 2 contained a novel transcript, Cdc45l2, that is strongly conserved between human and mouse. This gene is homologous at the amino acid level to three sequences from evolutionary distant organisms (yeast Cdc45p, Tsd2 protein from the smut fungus, U. maydis, and a C. elegans hypothetical protein F34D10.2) at least two of which are essential for DNA replication. The CDC45 mutant exhibits high rates of chromosome loss/recombination and accumulates chromosome damage. Experiments in yeast suggest that DNA replication involves an origin of replication complex (ORC) that is bound to replication origins throughout the cell (Owens et al. 1997). Shortly after mitosis, additional proteins join ORC to form a pre-replication complex (Pre-RC). Candidate proteins include Cdc6p and members of the MCM (mini-chromosomemaintainence) family (Owens et al. 1997). Cdc45p also appears to complex with the Pre-RC late in G1 (Aparicio et al. 1997). Initiation of replication requires two kinases, Cdc7p and Cdc28p in association with their respective regulatory subunits, DbF4p and CLb5p/Clb6p. Cdc45p and Cdc7p kinases are dependent on each other for the execution of replicative function (Owens et al. 1997). Once replication is initiated, the pre-RC disassociates and Cdc45p and the MCM proteins appear to move with the replicative fork (Aparicio et al. 1997). Tsd2p is structurally related to Cdc45p and is known to be required for DNA replication in U. maydis (Onel and Holloman 1997). Although it is not known whether CDC45L2 is a functional mammalian homolog of Cdc45p, it is possible that this protein is also involved in DNA replication.
The etiology of DGS involves a specific developmental pathway, namely, neural crest cell function (Budarf and Emanuel 1997), and contrasts with a putative role for CDCL452 in DNA replication. However, further study of CDC45L2 is required to determine if haploinsufficiency of this gene contributes to the chromosome 22 deletion syndrome phenotype.
METHODS
cDNA Selection
cDNA selection was performed essentially as described (Korn et al. 1992; Chadwick et al. 1997) with two complementary oligonucleotides (CDRI-A: 5′-GATCGAATTCACTCGAGCAT-3′, CDRI-B: 5′-TGATGCTCGAGTGAATTCGATC-3′). Amplified cDNA was digested withEcoRI restriction enzyme and ligated to phosphatasedEcoRI-digested pBluescript SK+ plasmid (Stratagene).
Conservation cDNA Selection
Conservation cDNA selection was performed with poly(A)+ RNA from 13.5 dpc and 14.5 dpc mouse embryos by standard methods with the following modifications (Korn et al. 1992; Chadwick et al. 1997). The cosmids/fosmids were prehybridized with 125-fold weight excess of both human Cot1 DNA (GIBCO BRL) and mouse Cot1 DNA (GIBCO BRL). The first round selection was washed at 65°C for 10 min at 2× SSC. In the second round of selection, the biotinylated hybridization products were divided between two eppendorfs: The products in one tube were washed at 65°C for 10 min at 2× SSC, whereas the others were washed at 65°C for 10 mins at 2× SSC and 0.2× SSC. The cDNA was eluted with a final amplification by PCR for 15 cycles. The cDNA products from the 2× SSC wash were called LS (low stringency) products and those from the 0.2× SSC wash were termed HS (high stringency) products.
Characterization of cDNAs
cDNA select subclones were picked into 96-well microtitre plates before spotting onto Hybond-N membranes (Amersham), and processed for colony hybridization. A PCR product for each individual cDNA selection clone was generated by vector primers, electrophoresed on a 1% agarose gel, and alkali blotted according to the manufacturer’s instructions (Pall Biodyne). Southern blots and Northern blots were hybridized by standard methods (Sambrook et al. 1989; Clontech). The probes used as positive controls in the experiments were either lab generated or came from a variety of sources: probes ES2, CTP, UFD1L, and CLTD were gifts from A. Baldini (Baylor College of Medicine, Houston, TX); J. Groffen (Children’s Hospital of Los Angeles, California); A. Pizzuti (Istituto di Clinica Neurologica, Centro Dino Ferrari, Universita di Milano, Italy); and H. Sirotkin (Albert Einstein College of Medicine, Bronx, NY), respectively, and TMVCF was an IMAGE clone (GenBank accession no.R55031). A 10-dpc mouse embryonic cDNA library (Novagen) was plated and screened by standard methodology. Human CDC45L2 ESTs were obtained from American Tissue Type Collection (ATTC). cDNA selection clones were sequenced on an ABI 373A automated DNA sequencer.CDC45L2 and Cdc45l2 cDNA clones were sequenced by a mixture of automated sequencing and manually by dideoxy chain termination method and primer walking. The DNA sequence was analyzed by programs available at the Human Genome Mapping Programme Resource Centre (HGMP-RC).
Acknowledgments
We are grateful to Brian Chadwick for his most helpful advice on direct selection, and to Gaetan Lee for the PCR analysis ofGscl expression. The Fugu cosmid library was supplied by the Human Genome Mapping Project Resource Centre (UK). The work was funded by The Wellcome Trust, The British Heart Foundation, and the Birth Defects Foundation.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.