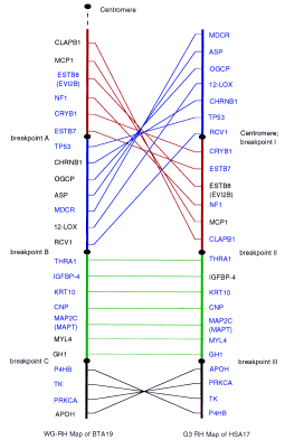
Parallel RH maps of BTA19 and HSA17. (Blue) Framework markers. The comparative RH map was divided into four regions with three breakpoints on each chromosome. Not only are the positions of region I (red) and region II (blue) inverted between human and bovine, but the gene orders within the two regions are also shuffled. (Green) Region III has the same gene order between human and cattle. The gene orders in region IV (black) between human and cattle are inverted.











