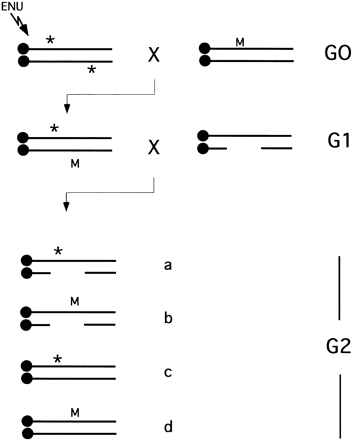
Region-specific saturation mutagenesis using ENU and deletions. Mutagenized mice (G0) with ENU-induced point mutations (*) are mated with female mice that harbor a dominant visible marker (M) and preferably a chromosomal inversion of the target region. The dominant marker allows tracking of the ENU-mutagenized chromosome and the inversion prevents recombination between the mutagenized and nonmutagenized chromosome. The offspring carrying a copy of the ENU mutagenized genome and the marker/inversion chromosome (G1) are mated with deletion heterozygotes. Four classes of (G2) progeny are generated in this cross: (a) Mice with the deletion in trans to the mutagenized chromosome; (b) mice with the deletion in trans to the marker/inversion chromosome; (c) mice that are heterozygous for the mutagenized chromosome; (d) mice heterozygous for the marker/inversion chromosome. Mice in class a are observed for novel recessive mutations, whereas the absence of this class indicates the induction of a recessive lethal mutation within the target region. In this case, G1 parent and/or class c siblings can be used to recover novel lethal mutation.











