WebWise: Guide to the Washington University Center for Genetics in Medicine Web Site
This installment of the WebWise series reviews the Washington University Center for Genetics in Medicine (CGM) web site. As with the previous WebWise reviews, you may find it useful to point your Browser Window to the CGM web site while reading. Redundant links are omitted from the site map displayed in Figure 1. The main features of this web site, as well as the features of those sites already reviewed, are indicated in Table 1.
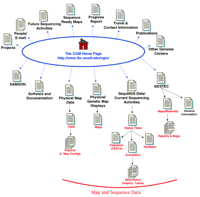
Site map for the Washington University Center for Genetics in Medicine. The main links to pages discussed in the text are illustrated here. Links on the top of the Home page are to general informational resources; links located on the bottom portion are to the data and additional tools. Some links available on the web site are not indicated here.
Features of the CGM Web Site
General Information
The CGM Home page utilizes the current “style standard” for informational web site presentation. A navigational toolbar is provided at the top of the page, additional links to more specific subject areas are provided along the left side of the page, and some introductory text is located in the body of the page. This general design is widely used by informational web sites as it facilitates navigation and information retrieval. The CGM web site presents this format on a few pages, but unfortunately this style is not used consistently on all pages. Although the navigation toolbar is only available on a couple of the internal pages, a Return to Top link to the Home page is supplied at the bottom of some (but not all) of the pages lacking the navigation toolbar. For pages without any navigation links, the bookmarked Home page, or the Browsers Go Back function allows one to return to a page of interest.
Several Home page links lead to general information about the CGM sequencing center. One can access the organizational overview via the toolbar Projects link, a more detailed description of the organization and project goals is available via the GESTEC link, and the CGM 1997 Progress Report via the Progress Report link. The GESTEC page, which is displayed in a new Browser Window, also includes a link to Maps/Materials documentation. From the Maps/Materials page (http://www.ibc.wustl.edu/cgm/searchs.html) one can access tables of STS markers, clones, CGM clone libraries, database descriptions, as well as a small amount of map and sequence annotation information. Postscript Map files can be downloaded from the SEGMAP examples page, and two annotation examples, available as either postscript or tiff files, can be downloaded from the Sequencing Analysis Examples page; it is not clear how up to date these files are. An overview of the CGM center’s longer-range goals is available by following the Future Sequencing Activities link. Contact information, including names, phone numbers, and e-mail addresses, can be obtained by following either the toolbar People link or the side column E-mail Contacts link on the Home page. These two links actually point to the same page and represent an unnecessary redundancy. By following the Travel and Contact Information link, one is led to maps and location information; and, as expected, the Publications link leads to a reference list, and the Other Genome Centers page presents a useful list of links to related web sites.
Data
The CGM sequencing center is working on mapping and sequencing several regions of the X chromosome. On the Sequence Data web page, the center reports having accumulated >6 Mb of X chromosome sequence data. Sequencing is carried out in collaboration with the Advanced Center for Genome Technology at Applied Biosystems Division of Perkin Elmer Corporation. The CGM web site presents both physical/genetic map data and sequence data. In addition to the downloadable maps available from the GESTEC page, the Home page includes toolbar and column links to map displays. Following the toolbar Physical Map Data link brings you to a page listing several report options such as reviewing lists of STS markers, clones, and map contigs. Following the Map Contigs link calls up a table indicating the region, description, quality (i.e., status), and contig name. The linked contig name brings up map displays of the region as a Postscript or Adobe Acrobat image. To review multiple map displays, reselect the file type before clicking on each subsequent link. If file type is not reselected, or if you do not have a PDF or Postscript viewer, then a download menu will appear. Although it is not as convenient to constantly reindicate the file type, one can take advantage of this system and elect to download a file by simply not reselecting the file type, or by selecting the wrong file type. Additional Adobe Acrobat displays of the entire X chromosome with two alternative labeling systems are accessible via the side column Physical/Genetic Map Displays links on the Home page. These large maps are displayed with a Java Applet on a Frames-formatted page, so make sure you enable Java in your Browser’s preferences options.
Surprisingly, the Sequence Ready Maps link leads to an outdated tabular summary of the sequencing targets rather than to the expected clone tiling path maps. Indeed, a map of the clone tiling path is not available on this web site. One must extrapolate the relative order information of the sequence data using provided marker data.
The Home page provides two links to the sequence data; namely, the toolbar Sequence Data link and the column Current Sequencing Activities link. Both links lead to a Status Table. These tables of data have a similar appearance yet provide some different data. It is not clear why two pages of sequence data are presented. Notably, the Sequence Data page reached via the toolbar link (http://www.ibc.wustl.edu/mod_perl/cgm_projects.cgi) lists more clones than the other data table. The page reached via the column link provides, albeit inconsistently, a more explicit indication of clones for which additional annotation data are available. Because the toolbar link leads to a table containing more data, that page appears to be more useful. Furthermore, accessing the Current Sequencing Activities page, (http://www.ibc.wustl.edu/cgm_SEQ/) via the column link is more cumbersome as you must first traverse an intermediate page where registration is requested. Contrary to appearances, registration is not required and one can simply follow the link to the data. In any event, this adds an unnecessary navigation burden and this intermediate page should simply be removed and the two tables of data consolidated into one.
Neither sequence data table includes a table legend, but the column headings provide adequate definitions for the most part. Both tables include information on location, clone name, sequence status, markers, insert size, and amount sequenced (the toolbar-linked table indicates larger numbers here). Links are provided in the Clone Type, Annotation, and Status/Genbank Accession columns. Note that the Annotation column is not included in the toolbar-linked table. One would expect that links provided in any given column would lead to the same type of information but alas this is not the case for the Clone Type column. These links lead to at least three different places on both tables: (1) to a page displaying the sequence in FASTA format; (2) to a directory listing of sequence files; or (3) to an Annotation page. There is no explicit indication of what type of information is to be obtained by following a given Clone link. This is an unfortunate policy that leads to confusion, limits the utility and accessibility of the data, and is generally bothersome.
Annotation is provided for many of the clones listed in both sequence data tables. Different amounts of information are provided, with the minimum consisting of a table listing locations and types of repetitive sequence identified in a given contig. Follow the bWXD42 clone link from the Sequence Data page (toolbar-linked) to review an example of the more detailed annotation provided for a known gene (http://www.ibc.wustl.edu/cgm_SEQ/bWXD42/index.html). This annotation page includes links to a description, the sequence, sequence composition information, a graphic representation, and an alignment. The description provided with each annotation includes a brief description of the sequence, its length, the GenBank accession number (inconsistently hot linked), and an acknowledgment of the persons involved in generating the data. Many of the annotation pages include links to both FASTA and Flat File format views of the sequence data. This information is displayed via the web browser and can be saved to a local computer by using a web browser to save the page as a text file. The repetitive sequence and exon location data are presented as simple tables. And the GIF of sequence features indicates GC and CpG content, location and orientation of repeats, exons (and gene names), and locations of rare cutter restriction sites. Additional tables of information derived by analysis with gene prediction programs are accessible via the small yellow buttons preceding the prediction program names.
The CGM web site does not include links to download the sequence data. However, the data is available either directly or indirectly via the Clone name links, or the linked GenBank accession number. Because update dates are not indicated on the sequence Status Tables, it is unclear whether the sequence available through the Clone link differs from the submitted GenBank sequence. If a given Clone link brings up the Annotation page, then you must traverse an extra page before reaching the page that displays the sequence in FASTA format. Once you reach this page, you can use your browser’s functions to save the page as a text file.
Tools
The CGM web site includes documentation on three software tools. This web site does not include either a search tool or a BLAST server. The toolbar SAMSON link leads to documentation of the “Sequence Analysis Management Served Over Networks” software program (http://cgmsamson.wustl.edu/), developed collaboratively by the CGM and Perkin Elmer. Note that the toolbar link to this page from some of the internal pages, for example, from the Software and Documentation page, brings you to an intermediate page, which in turn contains a link to the SAMSON page. This page includes links to the abstract presented at the recent Cold Spring Harbor Genome Mapping, Sequencing, and Biology Meeting as well as links to an interactive demonstration of the program. This program provides sequencing project management support and tracks the sequencing process from the colony picking stage to reaction tracking. The availability of this software is unknown; download links are not provided on the web page. A feedback mail link is provided, so inquiries can be conveniently directed to the center.
Descriptions of the Consed and FPC programs are accessible on the Software and Documentation page (follow the Home page link;http://www.ibc.wustl.edu/CGM/doc/software_documentation.html). The Consed program supports viewing and editing sequence assemblies generated with the Phrap program. Extensive documentation is provided about the Consed program, including instructions on obtaining access (contact the center). The FPC program (FingerPrinted Contigs) supports the creation of contigs, views of markers associated with clones, and facilitates selection of a minimal tiling path series of clones. Although this program was developed elsewhere, the CGM web site redistributes extensive documentation for the FPC program. This documentation is in PDF format so you must have the Adobe Acrobat Reader program installed on your local computer. This program is free and can be downloaded athttp://www.adobe.com/prodindex/acrobat/alternate.html. Information is not provided on availability of the FPC program.
Conclusions
The CGM web site presents both map and DNA sequence data for the X chromosome. Several groups are involved in sequencing this chromosome, and to obtain the grand overview, a series of good map displays are quite important. Although the CGM web site does provide a lot of physical and genetic map data, it does not provide a clear indication of the sequencing clone tiling path. One must deduce this information by looking at the ordered list of clones on the sequence data tables; but it would be reassuring if this information was provided in a graphic map display. The DNA sequence data tables do provide some linkage to the map data by listing the marker limits and STS content of each clone or contig; however, for most people, this does not sufficiently facilitate pulling the map and sequence data together. The DNA sequence data themselves must be tightly and conveniently correlated to the map data in the end; the map data tie it all together and provide the “big picture.” Ideally, one hopes to find a series of linked maps from a general cytogenetic display of the targeted region, to more detailed physical maps, to the maps of the tiling path of clones selected for sequencing. In the ideal situation, these latter maps would then be linked to the DNA sequence data. Taking full advantage of internet technology, the map and DNA sequence data should be accessed both as a linked series and as more direct links.
The overall ease of navigation at the CGM web site is rather uneven. The pages do not present a consistent design format or navigation links, and some pages appear to be redundant. Some links present the next page in the current Browser Window, whereas other links call up a new browser window when there is no obvious need for one. The registration page one must visit before getting to the Current Sequencing Activities page is unnecessary, and some of the general information pages have not been updated (at the time of this writing). Taken together, the ease of access and overall utility of this web site could be enhanced.
Next Month: University of Oklahoma’s Advanced Center for Genome Technology