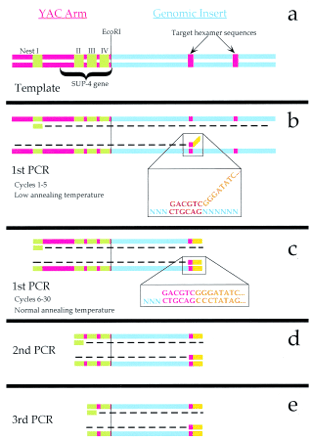
Schematic representation of vector–hexamer PCR. (a) The YAC vector/insert junction (right or left arm) before amplification. The YAC vector is shown in red (left) with nested vector priming sites in green. The first nested priming site is located outside theSUP4 gene. The mouse genomic insert is shown in blue (right) with two particular hexamer sequences shown in red. (b) The initial cycles of amplification at low annealing temperature. The vector primer (in green) anneals to its complement in the YAC arm, from which a new strand is synthesized (broken line). The hexamer primer is shown annealing to a target site in the mouse insert, its hexamer portion shown in red and its 20-bp anchor sequence in orange. The low annealing temperature (35°C) allows pairing and priming by the 3′ hexamer alone (shown in expanded view). (c) The later cycles of the first round of PCR are performed at a higher annealing temperature so that the entire hexamer primer stably anneals only to products from the initial cycles. (d,e) The second and third rounds of PCR reamplify the vector/insert junction products from the first PCR with nested vector primers to increase the specificity and yield of the amplification. The fourth nested vector primer allows reduction of vector arm contribution to the PCR product for use as a hybridization probe.











