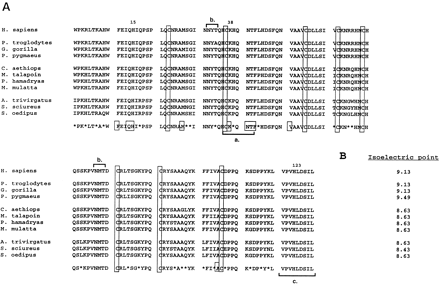
(A) Alignment of the primate orthologs of RNase k6. The DNA sequences encoding the polypeptides shown were generated by PCR from genomic DNAs as described. Each DNA sequence encodes an ORF that includes the eight canonical cysteines (shaded boxes), H15, H123, and K38 corresponding to residues found in the active site of RNase A (numbered), and the CKXXNTF motif (bracketa) common to all members of this superfamily. The amino acid sequence shown in the lowermost line denotes residues at which the eleven orthologs are identical (lettered) or divergent (*); residues included in boxes are common to all RNase A superfamily members (Riordan and D’Alessro 1997). Glycosylation sites initially identified in the human sequence are indicated by the brackets marked b.Each DNA sequence also encodes a 23 amino acid signal sequence (not shown); the residues demarcated by bracket c were included within the 3′ primer sequence. Common names for each genus/species include H. sapiens, human (GenBank accession no. U64998);P. troglodytes, chimpanzee (AF037081); G. gorilla,gorilla (AF037088); P. pygmaeus, orang-utan (AF037082);C. aethiops, African green monkey (AF037090); M. talapoin, talapoin (AF037087); P. hamadryas, baboon (AF037083); M. mulatta, rhesus monkey (AF037089); A. trivirgatus, owl monkey (AF037084); S. sciureus, squirrel monkey (AF037085); S. oedipus, cottontop tamarin (AF037086). The DNA sequence of human RNase k6 has been corrected from that originally reported (Rosenberg and Dyer 1996), resulting in the conversion of R53 to A. (B) pIs of the RNase k6 orthologs were calculated by the PEPTIDESORT algorithm of the Wisconsin Genetics Computer Group Program on-line at the National Institutes of Health.











