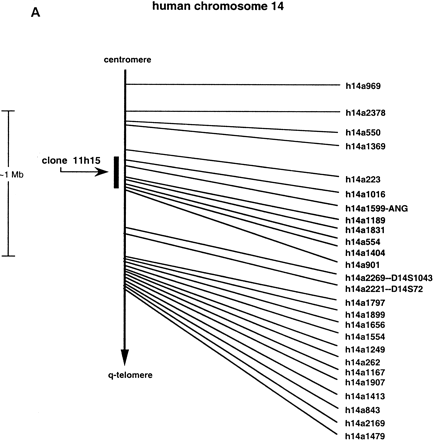
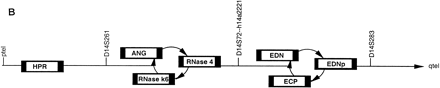
(A) Chromosomal localization of human RNase k6 by HAPPY mapping. Shown is a region of the HAPPY map of human chromosome 14 (Dear and Cook 1989, 1993; Walter et al. 1993; Dear 1997; Dear et al. 1997). The markers, STSs listed at the right are encoded by inter-Alu segments of genomic DNA; those located within one PAC distance of a previously defined marker (D14S1043, D14S72) are also noted. Clone 11h15 from the de Jong RPCI1 library (Ioannou and de Jong 1996) includes the gene encoding human RNase k6 and the five HAPPY map markers h14a1016, h14a1599, h14a1189, h14a1831, and h14a554, within a span that corresponds to the the q11 region of chromosome 14. Marker h14a1599 is found within the gene encoding angiogenin (marked ANG). The GenBank accession nos. for the STSs are as follows: h14a1016, G35789; h14a1599, G36057; h14a1189, G35876; h14a1831, G36118; h14a554, G35567. (B) Relative positions of the six human RNase A family members on chromosome 14. Assembling the information presented in Awith that available from the combined maps of the NCBI. (http://www.ncbi.nlm.nih.gov), the most likely linear arrangement of these ribonucleases can be inferred; those listed in circular fashion cannot be placed in a definitive order with respect to one another with the information available. (HPR) Human pancreatic ribonuclease/RNase 1; (ANG) angiogenin/RNase 5; (RNase 4) ribonuclease 4; (RNase k6) ribonuclease k6; (EDN) eosinophil-derived neurotoxin/RNase 2; (EDNp) eosinophil-derived neurotoxin pseudogene; (ECP) eosinophil cationic protein/RNase 3. The diagrams are not drawn to scale.











