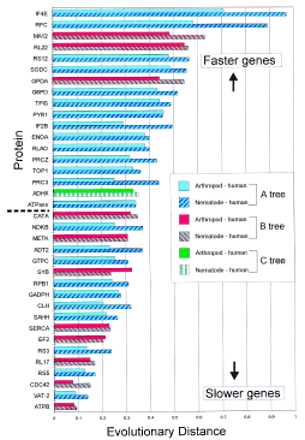
Relative evolutionary rates of the 36 protein quartets subjected to phylogenetic analysis. Protein quartets supporting tree A are shown in blue, those supporting tree B are shown in red, and the quartet supporting tree C is shown in green. The protein name abbreviation is shown along the y-axis, and the proteins are plotted in order of the mean evolutionary distance of nematode to human and arthropod to human where nematode is C. elegans, and arthropod is D. melanogaster. Proteins with the highest number of pairwise substitutions (fast evolving) are at the top and those with the lowest number of pairwise substitutions (slow evolving) are at the bottom. The evolutionary distances along the x-axis were determined from amino acid alignments using a Poisson correction as described in Methods. The broken line (middle left) represents the midway point where 18 proteins are above the line and 18 are below the line. The key to the bars is shown.











