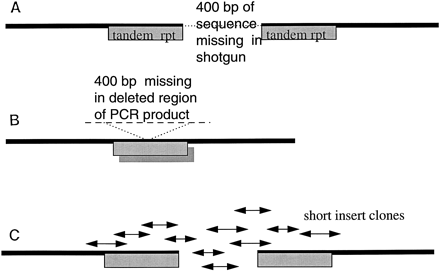
F59D12–C. elegans chromosome II. (A) Restriction digest revealed that although the assembly appeared contiguous, there was a 400-bp fragment missing between two identical repeat motifs. This was present in the cosmid but had become deleted from all shotgun subclones. (B) A PCR product was obtained across the region, but this concurred with the original deleted assembly. The PCR reaction had “skipped” between the two repeat regions giving a product that was also missing the 400-bp fragment. (C) A restriction fragment containing the missing sequence was isolated and sonicated to give a small insert library which, when sequenced, revealed the missing 400 bp. EMBL accession no. Z81558, bases 18910–19550. Sequence starts, GTCCACTTACGGGAAAAGGCAAAAATTTA; sequence ends, TTCCCATGACTTTCCGAAAAAAAGGCGGG.











