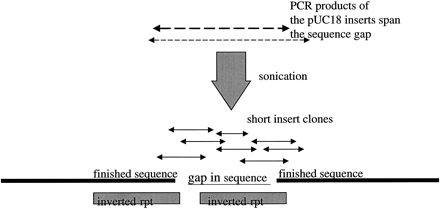
Y48E1–C. elegans chromosome II. Restriction digest analysis showed that a fragment of 800 bases was missing from the assembly, and although three pUC18 shotgun subclones spanned the gap, they were unsequenceable in that region. The small-insert clones obtained from the inserts of two of the shotgun pUC18 subclones provided complete and unequivocal contiguation of the gap, which could then be identified as containing one arm of a 1-kb inverted repeat. EMBL accession no.Z93392, bases 263250–264250. Sequence starts, ATCATGGTTGATAACGTAAATTCCCAGAC; sequence ends, CGCTGCGTATCGATTTTTATGAAACTGTG.











