WebWise: Guide to McDermott Center for Human Growth and Development at University of Texas Southwest Web Site
This, the seventh installment of the WebWise series, reviews the McDermott Center for Human Growth and Development at the University of Texas Southwest (UTSW) web site. Please note that the web address reported in the first WebWise installment (Pruitt 1997) has changed; the UTSW web site has relocated to http://gestec.swmed.edu/. As with all of the WebWise reviews, you may find it useful to point your Browser Window to the UTSW Home page while reading. This web site uses an abundance of links; to better illustrate the location of pages of interest, many links are omitted from the site map (Fig.1). The main features of this web site, as well as the features of those sites already reviewed, are indicated in Table1.
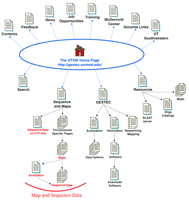
Site map for the McDermott Center for Human Growth and Development at UTSW. The main links to pages discussed in the text are illustrated here. Links on the top of the Home Page are to general informational resources, whereas the links located on the bottom portion are to the data and additional tools. Some links available on the web site are not indicated here.
Features of the UTSW Web Site
General Information
The overall browsing experience of the UTSW web site is enhanced by uniform style and organization presented on the pages. This web site emphasizes organization, navigation links, and a more sophisticated use of color, which has the overall effect of presenting a more polished web site. Pages are organized into categories, with links to these categories provided on the left side of the Home page (http://gestec.swmed.edu/). The organizational strategy used at this site is analogous to a branching tree structure—from the Home page several branches (main categories) can be reached from which additional branches (subcategories) emerge, and so forth.
Before beginning to browse this site it helps to have some understanding of how the navigation links are presented. The links located at the top-most portion of the Home page, which include the all-important link back to the Home page, are consistently available on all UTSW web pages checked, with the exception of the BLAST server page. A series of additional navigation links are presented on each page; their exact nature depends on the page at hand. In general, links relevant to the immediate subcategory are listed on the left side of each page, whereas links relevant to the last branch point occur on the top portion of each page. For example, when you follow the Resources link from the Home page, the navigation links that are displayed on the left side of the Home page are relocated to the top of the Resources page (header links). New navigation links that are specific to the Resources category are presented along the left side of the page (column links). As you delve deeper into the series of pages devoted to any given category you will always see the column links from the previous pages’ presented as header links on the new page, with a set of more detailed column links appearing. This approach to navigation is initially somewhat confusing until you become familiar with the general format, but it does provide very flexible navigation options. Furthermore, a visual cue indicates which link you just followed (e.g., which subcategory you are in)—a different graphic icon is used to indicate the link just followed.
The majority of the column links provided on the Home page lead to general information resources (these links are depicted on the top portion of the site map illustrated in Fig. 1). Contact information is provided on the bottom of the Home page, and each internal page provides a mail link to the UTSW webmaster. The informational links bring you to descriptions of the UTSW center, job and training opportunities, resource information (such as a catalog of oligonucleotides), as well as pages listing links to other web sites of interest. From the Home page, follow the McDermott Center link, and then the Maps and Views link if you need directions and maps (http://gestec.swmed.edu/maps&.htm). The Contents page (follow the top-most Contents link) brings you to a very useful overview of the web site that lists links to all of the UTSW web pages in an intuitive order; use this list to navigate directly to a more internal page. Some of the general information pages have very little content. For example most of the pages available from the Resources page are still under development, and the News page provides a link to a downloadable newsletter but little else. Because this web site recently has moved to a new address and undergone restructuring, it is understandable that some pages might still be “under development”; however, in such cases, it would be more beneficial to the user if those links were simply omitted for the time being.
Data
Follow the Sequence and Maps link from the Home page to access the data. The UTSW genome center’s main sequencing effort focuses on regions of chromosome 11 and 15, and a list of the sequencing targets is provided at the bottom of the Sequence and Maps page. Although these web pages do not provide an overview of the amount of DNA sequence data generated by the UTSW sequencing group, after perusing the sequencing pages it is apparent that this group is sequencing a sizable portion of chromosome 11 and ∼8 Mb of chromosome 15. Furthermore, the genome center indicates a commitment to sequencing all of chromosome 15.
The Sequence and Maps page (http://gestec.swmed.edu/sequence.htm) presents some general information about the UTSW data release policy (data are released daily) and provides, in the second paragraph, a link to the FTP site (ftp://mcdermott.swmed.edu/pub/SEQDROP/). Unfortunately, the file-naming system used on the FTP site does not impart any meaningful information about the sequence, such as the chromosome or specific target region it is derived from; hence, it is challenging to determine what is what. Data are accessed from the Sequence and Maps page via the column links. These links lead you to Chromosome 11 and 15 pages, to a Miscellaneous Human page, and to pages on other organisms. The Miscellaneous Human page only provides links to the sequence data. A map display or general description is not available for these clones, but contact information is provided so one can seek further information. The Chromosome 15 link will bring you to a page describing the medical significance of the sequencing effort in the 15q11 and 15q26 regions. The Chromosome 11 page presents a description of project goals and technical strategies. All three of these subcategory pages provide column links that focus in on the data for a specific target or clone. Navigation through this web site has been straightforward up to this point, as links are supplied in a consistent manner using meaningful nomenclature. However, as you enter the next layer of this web site, navigation becomes more cumbersome. To maneuver from the Home page to the Chromosome 11 page you must traverse three pages. Two to three more pages must be navigated before reaching the DNA sequence data. But for each individual target on chromosome 11 or 15 (8 and 2, respectively) you must successively move up and down two to three layers of pages to review the map and sequence data for the project as a whole. Furthermore, the link nomenclature imparts insufficient meaning on these inner pages. The links denote the cytogenetic region or clone name. It would be more useful if there was also some indication that a given link brings one to a map, to an annotation figure, or directly to the DNA sequence.
The column links on the Chromosome 11 and 15 pages lead, for the most part, to a map display of a given cytogenetic target. The chromosome 11 project consists of eight apparently noncontiguous targets scattered along the chromosome, and two targets are indicated for chromosome 15. Follow the column links provided for each chromosome-specific target to view the sequencing map. Map displays are not available for all targets, and in those cases by following the cytogenetic link (e.g., 11q23.3) you will be led to a page that includes links to the DNA sequence pages and little else.
The maps themselves use a fairly uniform style that indicates the clone tiling path and the general sequencing status (from Mapped to Finished); this important feature expedites interpretation of maps from different regions.
Most of the maps fit within the Browser Window or require minimal horizontal scrolling to view the whole map. In contrast, the 11p15.5 map is quite wide and one must scroll both horizontally and vertically to review the whole map. This makes it quite challenging to get the “big picture” or even to print out a copy of the map. Furthermore, the awkward effect of the background retiling is visible as you scroll (e.g., the purple side column reappears as you scroll to the right). On the plus side, one nice property of these maps is the inclusion of a color key on each map. Although the majority of the maps are static (e.g., you cannot click on them to jump to the sequence data), some (e.g., the 11p11.2 and 11q23.1 maps) are image-mapped, which directly connects the map and sequence data. Unfortunately, these image maps provide links to the sequence data for only a few of the clones and do not include any visual indication of status. All of the map display pages include column links that lead to either annotation data (discussed below) or to a display of the sequence data (e.g., the annotation page is not available for all clones). The link nomenclature reflects the clone name, and for the most part the clones listed on the side column can be found in the map display. However some exceptions were observed. For example, although a link to the sequence of clone pDJ205d23 is provided in the side column of the 11q23.1 map display page (http://gestec.swmed.edu/chromoso3.htm), this clone is not indicated on the map itself. The opposite was also observed on some maps, that is, some clones that are color-coded as finished on the map are not listed in the column links. The incomplete integration of the map and sequence data is confusing and diminishes the overall utility of the data.
The column links provided on the map display pages lead, for the most part, to DNA sequence data. The DNA sequence is displayed in FASTA format on a regular web page using the standard UTSW style (e.g., header links and background). The data on these pages cannot be downloaded directly; however, by using your computer’s “cut and paste” options you can save a local copy. Although explanatory text and/or an indication of length is not provided, the initial FASTA line (“>text”) does indicate the accession number for submitted sequences so one can look up additional information if needed. Sequence data are displayed as concatenated contigs for clones still being sequenced. Surprisingly, links are not provided to the relevant file in the public databases or on the UTSW FTP site.
A handful of the column links included on map display pages lead to annotation results rather than to sequence data (links to the sequence data are provided on the annotation pages). Annotation pages are provided for some clones in the 11p15.5, 11p11.2, and 15q26 targets. For example, follow the STR Cosmids link from the 11p15.5 map page (http://gestec.swmed.edu/str.htm). Annotation is provided as a graphic rendition at the top of the page, with tables of results under that. The graphic display indicates identified homologies to repeat elements, ESTs, other non-EST sequences, as well as predicted exons. Although a color key is provided, it does not include an explicit indication of scale—evidently the enumerated tick marks indicate each kilobase. Overall, the graphic display provides valuable information in a clear, concise manner. In contrast, the results provided in text form following the graphic are not as intuitive. The EST and non-EST GenBank homologies are summarized, respectively, by region; apparently the sequence is divided into noncontiguous regions that are analyzed individually for homology to ESTs or to non-EST GenBank sequences. Integrating these two results is more difficult than need be because different regions are defined for the two analyses. Overall, the correspondence between the figure and the text results could be improved by (1) indicating the analysis regions on the graphic display, (2) using the same region definition for the two homology-based analyses, and (3) linking results to the GenBank record. Nevertheless, this type of overview of DNA sequence data can be quite useful to “gene hunters” and the research community in general.
Tools
The UTSW web site includes the two tools most essential to a sequencing center web site—namely, a search tool and a BLAST server. This center also provides concise descriptions of automation efforts and software tools and, notably, obvious links to download the tools described. The search tool is accessible from every web page on the top-most set of navigation links and can be used to find web pages, maps, and sequences. After typing in a key word and starting the search, all results meeting the query are returned in a table format. For the most part, the document titles indicated are sufficiently descriptive to allow identification of the page of interest. Overall, the search tool is easy to use, performs well, and returns useful results.
Follow the Resources link from the Home page to the BLAST server link to access the UTSW BLAST server (http://gestec.swmed.edu/blast.htm). At the time of this writing, only some of the UTSW sequence data, namely that of chromosome 11 clone-end sequences, are included in the UTSW BLAST database. The web page indicates that a more comprehensive database will soon be available. The web sites of sequencing centers’ provide a critical public resource, and it is important to include access to the complete sequence database. The BLAST server page itself departs from the standard UTSW web format in that it presents a gray background and no navigation links. To submit a query simply paste your sequence of interest into the text box, in FASTA format but without the “>comment” line, and click on the Perform Search button. Although results are returned very quickly, the ambiguous hit descriptions and lack of links to either the UTSW sequence data or to the public databases limits this tool’s utility.
Follow the GESTEC link from the Home page to access descriptions of automation strategies and software tools. Some of the pages reached via the GESTEC page contain very little content, such as the Sequencing and Chromosome 11 mapping pages. From the GESTEC page, follow the Automation to the Projects link to access the UTSW automation strategy. Unfortunately the Projects page merely presents a new list of links instead of also including a general overview. Following the provided links brings up a page that includes an illustration of the equipment as well as a description. In addition, some relevant bibliographic information is available via the Resources link on the GESTEC page. Descriptions of software tools are available from the GESTEC page by following the Informatics link. This brings you to a page that includes several short, concise descriptions of the software. Although extensive documentation is not available here, one pleasant feature of this web page is the inclusion of contact information and clearly labeled download links.
Conclusions
The UTSW sequencing center presents a polished web site with emphasis on organization and convenient navigation. This web site includes the essential information of map and DNA sequence data, descriptions of software tools (with clear download links), a useful search tool, and a BLAST server. Additionally, supplementary data for some clones are provided in the form of graphic and text annotation. Annotation of high-throughput sequence data via a web interface makes sense. The DNA sequence by itself imparts no meaningful information; the map data provide an overview of the location of the sequence data, and perhaps some correlation to a disorder of interest. But annotation, as either indications of homologies or open reading frame predictions, is a prerequisite to gene identification. As discussed in the previous WebWise review (Pruitt 1998). The method of annotating sequences is somewhat controversial. Providing annotation via the web is an obvious solution provided the annotation is periodically updated.
Organization is a crucial component of effective web design, but the inner pages of this web site are subject to excessive organization. Although the organization and provision of useful navigation links does allow convenient movement through the “top” levels of this web site, maneuvering through the inner pages is cumbersome. The inner pages are divided into a profusion of subcategories, and the type of data a given link leads to is not always obvious. Furthermore, the link style adopted here is initially confusing. The current state of the art for designing informational web sites is to provide a set of consistent web site navigation links at the top of each page and to include more specific subcategory links along the side or bottom of the page. A uniform set of column links are then displayed on each page relevant to a given subcategory, and if needed, more specific links can be provided in the body of the page. In addition, distinct styles are typically used to visually separate the header and column links. As currently designed, the UTSW pages can be slow to download and appear somewhat cluttered. Some minor link restructuring and reorganization of the map, annotation, and sequence data for each chromosome would improve data access. Tufte (1997) recommends a simpler, more streamlined approach to data presentation; this approach is also recommended for web site design in general (Siegel 1997).
Nonetheless, the UTSW web site does have the essential elements in place, and there are indications of future intentions that will enhance overall access to, and utility of, the data. For instance, some of the maps are already image-mapped; the continuation of this trend will make all of the data more useful by integrating sequence and map data more tightly. In general, providing image maps is a very effective, intuitive way to link these two types of data. Additionally, this web site indicates that the BLAST server will soon include all of the UTSW sequence data, which will enhance the overall utility of this important tool.
Footnotes
-
↵1 E-MAIL ; FAX (301)-435-2433.
-
Next month: University of Washington Genome Center
- Cold Spring Harbor Laboratory Press