WebWise: Guide to the Stanford Human Genome Center and the Whitehead/MIT Genome Center Web Sites
This installment of the WebWise series reviews two web sites: (1) the Stanford Human Genome Center (SHGC;http://www-shgc.stanford.edu/), and (2) the Whitehead/MIT Genome Center (W/MIT; http://www-genome.wi.mit.edu/). Although the overall organization and style of these two web sites are quite different, the centers share an important feature. Both sites make available a large amount of physical map data and supportive mapping-related resources. Although these data and resources are extremely valuable to the research community, the objective of this series is to highlight the data and resources pertaining to the sequencing effort; therefore the physical mapping component of these web sites will not be covered in detail here. When comparing these web sites to the site maps depicted in Figures 1 and 2 (below), you may notice that some of the internal physical map-related links have been omitted from the site. In addition, many duplicate and minor links have also been omitted from Figures 1 and 2. The main features of these web sites, as well as the features of those sites already reviewed, are indicated in Table1.
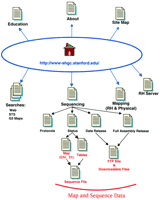
The Stanford Human Genome Center site map. The main links to pages discussed in the text are illustrated here. Links on the top of the Home page are to general informational resources, whereas the links located on the bottom portion are to the data or additional tools.
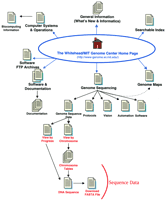
The W/MIT Genome Center site map. The main links to pages discussed in the text are illustrated here. Links on the top of the Home page are to general informational resources; links located on the bottom portion are to the data or addition.
Features of the SHGC and W/MIT Genome Center Web Sites
The SHGC
The SHGC has sequenced ∼7.5 Mb of chromosomes 4 and 21 and has targeted an additional 10 Mb on these chromosomes. In addition, SHGC has designed a simple and concise web site to make this and other information publicly accessible. This web site is nicely organized, and navigation between pages is facilitated by inclusion of useful internal navigation links at the top of most pages. These internal links are quite handy and eliminate the odious task of hitting the Go Back button repeatedly should you want to jump over to a different general topic from an inner page.
General Information
The links to more general, descriptive information are depicted on the top portion of the site map (Fig. 1). General information, including contact information and useful links, is available from the About page. The Staff list, although certainly useful, does not clearly specify an individual to contact for a particular type of question. Many web sites clearly identify the contact point for questions about the web site, or for questions concerning sequence analysis. A fairly comprehensive list of Human Genome Project web sites is available on the Links page. A list of many general biology and science web sites is also available on the Resources page (follow the Education, and then the Resources, link). One very useful feature included on this web site is the Site Map. The Site Map, accessed by following the text link at the bottom of the Home page, lists all of the pages and links available on this web site. It is organized in a manner to reflect the overall organization of the web site and includes links to all of the internal pages. This can be quite useful if you are looking for a particular inner page and wish to avoid jumping through several intermediate pages.
Data
To access the sequence data, follow the Sequencing link from the Home page (see Fig. 1). From this page (http://www-shgc.stanford.edu/Seq/index.html) you can access Sequencing Protocols, Sequencing Status, and two data download pages. The Sequencing Protocols page provides some basic laboratory protocols such as DNA preparation and sequencing reactions. Disappointingly, it does not include any information on the sequencing software or data management tools being developed and/or used by the SHGC.
The SHGC has set a goal to sequence 17.5 Mb of human DNA over 3 years. It has begun this initiative by targeting ∼7.5 Mb of chromosomes 4 and 21 for sequencing. Follow the Sequencing Status link from the main Sequencing page to see an overview of the main targets (http://www-shgc.stanford.edu/Seq/Status/index.html). Additional information about the three primary targets (the Down Syndrome Critical Region and the EPM1/APECED region on chromosome 21, and the chromosome 4q25 region) is available by following the relevant link. Unfortunately there is some inconsistency in the presentation of sequencing data; an image map is provided for one target and tables are provided for the remaining targets. Sequencing status for the EPM1 and 4q25 targets is available in a tabular form that lists the clone name, hot linked to a sequence record, and the sequencing status. The sequence records called up through these links are formatted as they might appear in the public databases; links are not provided to the equivalent records in the public databases. Unfortunately, there is no indication of the relative clone order in the tables. The sequence data are much more useful when some indication of relative order, such as a map, is available. An image map, which depicts clone names, tiling path, and sequencing status, is only available for the 0.4-Mb Down’s Syndrome Critical Region of chromosome 21. One valuable feature of this map is the grouping of the clones into three contigs (DSCR_Left, DSCR_Middle, DSCR_Right); clicking on a clone or contig calls up a sequence file of the relevant contig. For this group of clones the separate clone sequences are available on the FTP site and a file that joins the three contigs illustrated on the image map is available from the Preliminary Full Assembly Download page. One potentially confusing discrepancy was noted between the image map and the contig sequence files; the initial letter of the clone names changed. Clones included on the image map all began with the letter A, whereas those listed in the sequence file all began with the letter Q (but were otherwise the same as the image map names). This is undoubtedly a reflection of internal record keeping but is a potentially confusing practice.
DNA sequence data are made available in several formats. As mentioned above, sequences are presented in a familiar sequence record format from the Sequencing Status pages. Data are also available to download or view on the FTP site as individual subclone reads or as preliminary assemblies of the subclone sequences for a given BAC clone. The smaller sequences are available via the 3Kb Clone Data Release link. A table lists the clone name, size, and origin, and links are provided to download data as a compressed file or to view the sequences available on the FTP site. The preliminary assembled sequences are available from the Preliminary Full Assembly Release page. These data files consist of a series of sequences in FASTA format. Files indicating sequence quality at each base position are also available for each sequence. These sequence files can be either viewed on your monitor or downloaded to your computer. To view the sequences, simply click on the appropriate icon. To download these FTP files to your local computer, hold down the shift key while clicking on the icon.
Tools
With a single exception, the SHGC has not included software tools or analysis capability on this web site. The SHGC web site does provide some search capabilities; links are provided on the Home page to search the web site or to search for STS markers or RH maps. The web search tool can be used to search for a web page or a sequenced clone name. This tool should be useful for anyone interested in monitoring the sequence progression of a given clone. Unfortunately, this web site does not include the ability to do a homology search. It is very useful to be able to carry out a BLAST search against a given center’s sequence data. Undoubtedly much of this sequence is already available through the public databases, but some may prefer to target a homology comparison to the originator of the data, where presumably you will have access to the most up-to-date sequence data.
Conclusions
The overall aesthetics of this web site are quite pleasing. The pages are simple, consistent, and easy to navigate. Although this is one of the smaller sequencing centers, with only 7.5 Mb currently under way, the SHGC has targeted the sequencing of an additional 10 Mb within 3 years. Sequence analysis and management of the resulting data have become increasingly dependent on software tools; in light of this, it is notable that the SHGC web site does not make any mention of the computational strategies that they employ for their sequencing, assembly, and data management tasks. Unfortunately (and surprisingly, given the large physical mapping focus of this group), image maps are not provided for each of the regions currently being sequenced. In general, image maps provide a very useful method to use to illustrate sequencing data as they successfully integrate several types of valuable information: clone names, the clone order (or tiling path), relevant chromosome markers, and convenient links to the actual DNA sequence. It enables one to quickly ascertain the general location of the sequence data, the relationship between the clones, and can conveniently connect the map data to the sequence data. On the plus side, the SHGC does provide the DNA sequence data in a variety of formats and also makes available information about the quality of each sequence.
The W/MIT Genome Center
The W/MIT Genome Center is in the process of sequencing all of chromosome 17 and regions of chromosomes 6, 9, 13, X, and Y. The Sequencing Center currently has ∼10 Mb of human DNA sequence, 7 Mb of which is from chromosome 17, available on their web site. The W/MIT Genome Center web site includes data and information originating from several groups (e.g., separate mapping and sequencing groups) and utilizes distinct styles for different sections of the web site. The Genome Sequencing link on the W/MIT Genome Center Home page (http://www-genome.wi.mit.edu/) brings you to the W/MIT Sequencing Center Home page (http://www-seq.wi.mit.edu/). Because some useful information, including software tools, is available from the more general W/MIT Home page, this is the starting point for the review and for the site map illustrated in Figure 2.
General Information
The main W/MIT Genome Center Home page presents a long list of links organized by general category. The site diagram shown in Figure 2omits many of these links, and in some cases a group of links is represented by a single arrow. For instance, the several links available under the What’s New category are represented by a single arrow on the site map. The What’s New category includes links to some information of general use (including contact information and directions), but it also includes links to pages intended for more internal use, such as usage statistics and seminar series. Contacting the webmaster through the mail link on the main Home page (WebMaster{at}genome.wi.mit.edu) proved to be a very efficient way to submit questions. The response was rapid and it was obvious that an effort was made to forward each question to the most appropriate person. Unfortunately, this useful contact information is not included on the Sequencing Center’s pages. The Computer Systems and Operations Home page link leads to a page listing additional contact information and computer help resources. Some of the links on this page have restricted access but are not labeled as such (e.g., Whitehead Institute Computing Group link). A very nice collection of links to other web sites is available by following the Biocomputing Information link. The Searchable Index tool, located at the bottom of the Home page, was not very useful for locating clones or markers, and for one of the searches tested (a marker name), returned a long list of unrelated items.
Follow the Genome Sequencing link to access the W/MIT Genome Sequencing Center Home page. These pages include navigation links at the bottom of each page and present a uniform style; however, the aesthetics could be improved somewhat if the heavy table borders seen on the data pages were removed. Follow the Vision link to obtain general information about the sequencing center, including staff information, and descriptions of finishing criteria and data release information. Several wet lab protocols are outlined on the Protocols page, which also includes an interesting review on formalizing naming strategies to facilitate data-tracking tasks. Note that the Help page link has restricted access, although it is not labeled as such.
Data
Follow the Genome Sequence Data link from the Genome Sequencing page to access the data. Overall sequence progress is indicated at the top of the page (in kb), but this number is not a very useful statistic as it reflects the amount of both human and mouse DNA sequenced. DNA sequence information can be viewed by progress or by chromosome. These two views contain the same data but organize it differently. The View by Progress page includes additional statistics at the top of the page, indicating the amount of sequence in different phases of completion. Clones on these pages are ordered first by status (finished clones listed first) and then by size (largest sequenced clones listed first). This has the effect, in the View by Progress page, of interspersing the human and mouse clones. Overall, the View by Progress page seems redundant and less useful than the View by Chromosome pages. To reach the Chromosome table (a summary) from the View by Chromosome page, one must traverse an extra mouse click, but the added organization of the data makes this step more than worthwhile.
A Chromosome table is provided for each chromosome (follow the appropriate links from the View by Chromosome page). This page includes an indication of the total kb of finished (and nearly finished) sequence, as well as a table of information about the clones. The table includes significant information, including two names for each clone (Clone name and Internal name), the names of any associated markers, the size (kb) of the sequence, the clone type (e.g., BAC), the status (e.g., finished), and links to the DNA sequence. It is not clear why two names are indicated for each clone, and one would anticipate that this practice could lead to considerable confusion. Unfortunately, there is no information concerning the relative order of the clones or whether any of the clones overlap. Some indication of clone order could likely be derived using the associated marker information, but this would be a time-consuming and tedious process. It would be very useful to have a graphic depiction of the sequence-ready maps with clone order indicated.
Links to the sequence data are provided for each clone in the tables. The sequence is available on standard html pages in FASTA format. The name used to identify each sequence is the Internal name and not the Clone name. Although it is undoubtedly a reflection of internal data management strategies, this practice invites confusion. Links are provided at the top of each sequence file to (1) the GenBank record, (2) download the sequence (in FASTA format) to your local computer, and (3) the Staden Package web site [(Staden 1996) the Staden Package is a sequence project management tool]. Downloading sequences was successful on both a PC and Sun computer; presumably downloading would also be successful on a Macintosh, but this was not tested.
Tools
The W/MIT web site does include a great deal of software documentation as well as descriptions of automation methods. The link to Automation and Development from the Genome Sequencing Home page leads to descriptions and pictures of seven robotic automation systems being developed by the W/MIT Sequencing Center. These systems range from automated plaque picking to DNA sequencing and appear to be currently under development; nonetheless, they represent an impressive package and future progress reports should be interesting. There is also a description of nine informatics tools being used and/or developed. These tools are described under the Software pages [follow the Software link from the Genome Sequencing Home page (http://www-seq.wi.mit.edu/informatics/)]. The software tools range from lane tracking, to quality assurance, to finishing. Availability is only made explicit for one tool, Trout Analysis (signal-processing and base-calling), which is available by anonymous FTP (genome.wi.mit.edu—in distribution/software/trout). Personal communication with the W/MIT Center via the WebMaster indicated that all software tools are available via FTP. Many items are found in the /distribution/software/ directory of this FTP site; however, it was not clear that all of the tools described in the web pages are actually included in this directory.
Additional information on software and development is available from the main W/MIT Genome Center Home page by following the Genome Center Software Documentation link (seehttp://www-genome.wi.mit.edu/genome_software/genome_software_index.html; Fig. 2, Software & Development). Two mapping-related tools are described here, as well as a data management tool (LabBase) and a primer prediction tool (Primer3), which can also be run via the web. LabBase, Primer3, several mapping programs, and Bass (lane-tracking and base-calling) can be downloaded by following the Genome Center ftp Archive link from the W/MIT Genome Center Home page (http://www-genome.wi.mit.edu/ftp/distribution/software/). This html page presents a very useful organization of the software available; each software tool listed is linked to the relevant FTP directory from which the programs can be downloaded. This approach works very well and is intuitive. The software tool pages accessible from the Genome Sequencing page would be enhanced if a central page listing each tool, and linking to the appropriate download site, were available.
Conclusions
The W/MIT Sequencing Center pages are easy to browse as they use a simple organization strategy, provide internal links to other pages, and use a similar design style. DNA sequence data are directly available via the web site, and links from the clone information to the sequence data are provided; however it is regrettable that there are no graphic depictions or other indications of clone order. The W/MIT Sequencing Center has engaged in sequencing an entire human chromosome, in addition to some smaller regions on several other chromosomes. This is a vast undertaking and the Sequencing Center is exploring numerous automation techniques, both software and robotic, to enhance sequence output. In light of this impressive investment in automation and software development, it is somewhat remarkable that there are not more tools available for direct use on the W/MIT Sequencing Center web site. The site does not include any web-based analysis tools, such as a BLAST server to carry out homology searches against the sequence data. Furthermore, there are no useful search tools available. It is very convenient to have the option to search the web site, or to search directly for the sequence data of a given clone or contig. On the plus side, the software tools developed are publicly available, which is a useful resource for the sequencing community.
Footnotes
-
↵1 E-MAIL ; FAX (301) 435-2433.
-
Next month: Baylor College of Medicine
- Cold Spring Harbor Laboratory Press