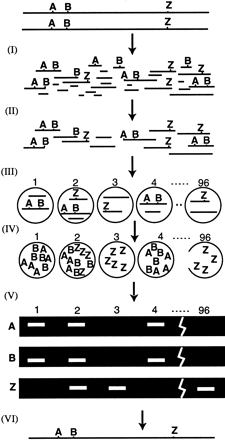
How HAPPY mapping works. Intact genomic DNA is broken by irradiation (I) to give a pool of random fragments, which are size selected by PFGE (II). A mapping panel of 96 aliquots is taken (III) from this pool. Each aliquot contains ∼1 haploid genome’s worth of DNA, so each marker is present in only a subset of the aliquots. (IV) The panel is pre-amplified a few hundred-fold by primer extension preamplification (PEP). (V) Subfractions of the pre-amplified panel are screened for specific markers using nested PCR. Markers A and B, (e.g., are found to cosegregate frequently. This implies that they tend to lie on the same fragment of DNA, and so must be tightly linked. A map may be constructed (VI) by examining cosegregation frequencies between all markers.











