
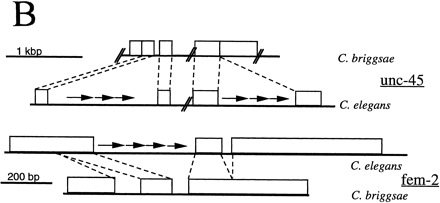
Arrangement of the CeRep25B repeat sequences within the fem-2and unc-45 genes. (A) Sequences of CeRep25B-containing intron sequences for C. elegans (Ce)fem-2 and unc-45, and the corresponding intron sequences from the related nematode Caenorhabditis briggsae(Cb). Intron sequences are in lowercase, exon sequences in uppercase letters. Translation is shown in single-letter code under the flanking exon sequences. CeRep25B sequences (both full repeats and half elements) are shown in bold. In the C. elegans fem-2 intron sequence, the portion deleted in a recovered E. coli clone is underlined dotted, and the endpoints of the deletions lie within the doubly underlined portion. In the C. briggsae unc-45 sequence, no intron is found in the corresponding position to Ce–unc-45intron 8, which is marked on the DNA sequence with an asterisk (*). (B) Schematic outline of the C. elegans and C. briggsae homologs. Arrows indicate the position (but not the number) of the CeRep25B sequences. CeRep25B sequences account for most of the difference in size of the introns.











