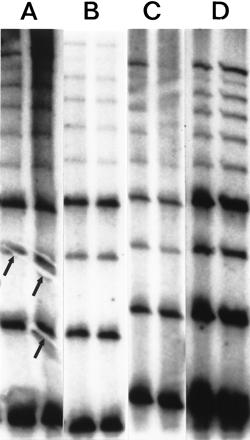
The effects of various blotting and hybridization techniques on RED of DNA from cell line GM 10850 using a (CTG)10 oligonucleotide. (A) Electrotransfer, 2 A for 40 min in 1× TBE. Prehybridization and hybridization in Amersham Rapid Hybe for 30 min and 45 min, respectively. The membrane was washed for 20 min at room temperature and for 2 × 15 min at 60°C in 1× SSC, 0.1% SDS. Typical electroblotting artifacts are shown (arrows). (B) Wet capillary gel transfer. The completed gel was placed on top of a stack of wet filter papers, overlaid with a sheet of wet membrane, covered with another sheet of wet filter paper, then topped with a stack of dry filter sheets and a weight for 2 hr. (C) The DNA was transferred from the gel onto a membrane by dry capillary blotting. A wet (1× TBE) Hybond N+ membrane was placed on the gel, overlaid with three dry 3M papers and a weight for 2 hr. Following UV immobilization the membrane was prehybridized for 30 min and hybridized for 45 min at 60°C in CHURCH solution. Thereafter the same washing procedure were used as in A and B. (D) Dry capillary blotting as described above in C. Hybridization and washing conditions as in A and B.











