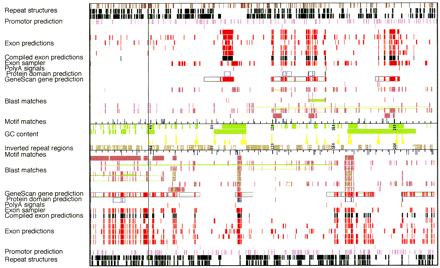
Graphic output of RUMMAGE results of the EPO contig. The vertical line represents the sequencing gap in the EPO contig. Rectangles define the regions in which matches were found. Corresponding matches are connected by green lines. Repeat structures are derived from Repeatmasker, Censor; exon predictions from GENESCAN, GRAIL2, FEXHB, MZEF, XPOUND; promotor prediction from ProScan; motif matches from DPS; poly(A) signals from Pol II; protein motifs from PROSITE; BLAST matches from BLASTS of various databases, Exon sampler. For references, see Methods. The graphic and tabular output of the automated first-pass annotation of both the EPO and theCUTL1 contig with RUMMAGE-DP is viewable via our Home page athttp://genome.imb-jena.de/.











