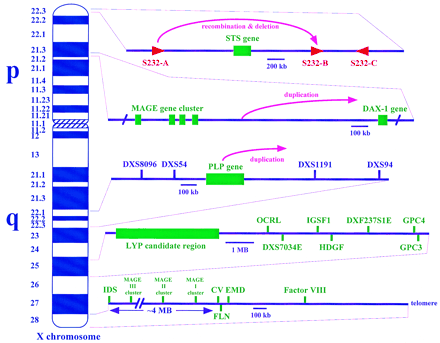
X chromosome regions with duplicated sequences involved in pathology. The regions are (from top to bottom) the X-linked ichthyosis region in Xp22.3, the dosage-sensitive sex reversal candidate region in Xp21.3, the Pelizaeus-Merzbacher region in Xq22, the lymphoproliferative syndrome in Xq25, and regions in Xq28 (further diagrammed in Fig. 2). The genes and probes indicated are all described in the Genome Database and include (STS) steroid sulfatase; (S232-A,B,D) three repetitive sequences that cross-hybridize with genomic clone CRI-S232; (MAGE) melanoma antigen gene family; (DAX-1), critical region for adrenal hypoplasia congenita; (PLP) proteolipid protein; (DXS8096, DXS54, DXS1191, and DXS94) polymorphic markers; (LYP) lymphoproliferative syndrome locus; (OCRL) oculocerbrorenal (Lowe) syndrome gene; (IGSF1) immunoglobulin superfamily gene 1; (HDGF) hepatoma-derived growth factor gene; (GPC3) glypican 3; (DXS7034E and DXF237S1E) expressed sequence tags; (GPC4) glypican 4; (IDS) iduronate-2-sulfatase gene; (CV) color vision genes; (FLN) filamin gene; (EMD) Emery-Dreifuss muscular dystrophy (emerin) gene; andfactor VIII gene.











