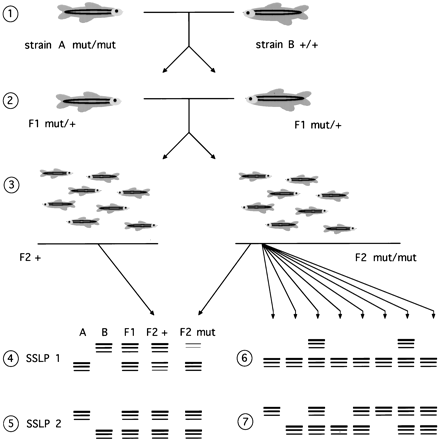
Linkage analysis using SSLP genotyping of pooled samples. (1) Fish of strain A homozygous for the recessive mutation mut are crossed to wild-type fish of strain B to generate F1 fish that are heterozygous at all polymorphic loci. (If homozygous fish cannot be bred, then F1 fish heterozygous for the mutation can be generated by mating a heterozygous mut/+ A parent with a wild-type B parent and identifying mutation carriers by progeny testing.) (2) F1 mut/+ fish are intercrossed and DNA is prepared from (3) phenotypically normal F2 progeny (which will be either wild type or heterozygous at mut) and mutant F2 progeny (which will be homozygous mut/mut). Samples from these two classes are pooled separately and tested using SSLP markers distributed across the genome. (4) For a marker linked to the mutation (SSLP 1), the PCR products corresponding to the A allele of the SSLP will predominate when the mutant pool is tested. (5) For unlinked markers (e.g., SSLP2), the abundance of PCR products of the two alleles amplified from the mutant population will be equal, and the distribution will appear to be the same as those from the F1 parents and the wild-type pool. (6) Linked markers are then tested in individual mutant progeny to determine their map positions with respect to the mutation. Two out of eight mutant progeny are heterozygous here. Because these are intercross progeny, this corresponds to 2 recombinants in 16 meioses, which is 12.5 ± 8.3 cM. (7) The distribution of SSLP 2 markers in individualmut/mut progeny is seen to be random.











