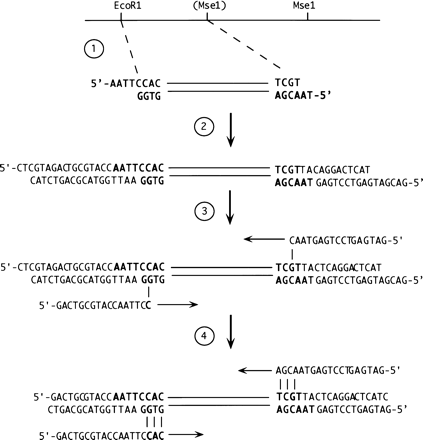
AFLP analysis (adapted from Vos et al. 1995). (1) Genomic DNA is digested with a combination of restriction enzymes. The products of this digestion will include some that are strain-specific due to sequence variation at the enzyme recognition sites (e.g., theMseI site shown in parenthesis). (2) The restriction products are ligated to adapters that carry cohesive ends for the respective enzymes and additional sequences to facilitate adapter-specific amplification. (3) A “preamplification” step is performed using primers corresponding to the adapters and each carrying an additional unique 3′-terminal nucleotide. This serves to limit the number of amplified products, as the template for this base will be derived from genomic sequence and will vary. Amplification of fragments carrying the same adapters (e.g., derived fromMseI–MseI restriction fragments) is less favorable than amplification of fragments with different ends, presumably because of the formation of stem–loop structures. (4) The population of products is further limited by amplification using a primer with three unique 3′-terminal nucleotides. These products can be separated on a denaturing gel and examined to identify the subset that are strain-specific. A kit containing reagents for AFLP analysis is commercially available from Life Technologies, Inc.











