Development of a Comparative Genetic Linkage Map for Armigeres subalbatus Using Aedes aegyptiRFLP Markers
Abstract
One of the causative agents of lympahtic filariasis is the nematode parasite Brugia malayi that requires a competent mosquito vector for its development and transmission. Armigeres subalbatus mosquitoes rapidly destroy invading B. malayimicrofilariae via a defense response known as melanotic encapsulation. We have constructed a genetic linkage map for this mosquito species using RFLP markers from Aedes aegypti. This heterologous approach was possible because of the conserved nature of the coding sequences used as markers and provided an experimental framework to evaluate the hypothesis that linkage and gene order are conserved between these mosquito species. Of the 56 Ae. aegypti markers tested, 77% hybridize to genomic DNA digests of Ar. subalbatus under stringent conditions, with 53% of these demonstrating strain-specific polymorphisms. Twenty-six Ae. aegypti markers have been mapped using an F2- segregatingAr. subalbatus population derived from a cross of strains originating in Japan and Malaysia. Linear order of these marker loci is highly conserved between the two species. Only 1 of these markers, LF92, was not linked in the manner predicted by the Ae. aegypti map. In addition, the autosomal sex-determination locus that occurs in linkage group 1 in Ae. aegypti resides in group 3 in Ar. subalbatus. The Ar. subalbatus map provides a basic genetic context that can be utilized in further genetic studies to clarify the genetic basis of parasite resistance in this mosquito and is a necessary precursor to the identification of genome regions that carry genes that determine the encapsulation phenotype.
[The composite map and sequence database information for Ae. aegypti markers can be retrieved directly from the Ae. aegypti Genome Database through the World Wide Web: http://klab.agsci.colostate.edu.]
Mosquito-transmitted diseases continue to cause significant human morbidity and mortality throughout the tropics and subtropics (TDR News 1994). Disease agents, including filarial worms and the protozoan species that cause malaria require a compatible mosquito host to complete their life cycles. Studies of vector competence have, until recently, relied on physiological and biochemical approaches and have provided limited understanding of the molecular events determining the success of specific mosquito–parasite interactions. A purely genetic approach could prove useful in identifying genes that confer the resistant phenotype in mosquito strains unable to transmit parasites. Genetic linkage maps recently have been constructed for two important disease vectors, Aedes aegypti and Anopheles gambiae, based on restriction fragment length polymorphism (RFLP) markers, microsatellite markers, and random amplified polymorhpic DNA (RAPD) markers (Severson et al. 1993; Antolin et al. 1996; Zheng et al. 1996). An advantage of the cDNA-based RFLP markers is that many of the sequences are conserved among mosquito species and, consequently, can be used for construction of comparative linkage maps (Severson et al. 1995a). A comparative linkage map for Aedes albopictus, based on Ae. aegypti markers, showed that linkage and gene order are completely conserved at 18 loci spanning all three linkage groups (Severson et al. 1995a).
The mosquito Armigeres subalbatus, mounts a rapid defense response against invading Brugia malayi microfilariae. This form of invertebrate immune response is known as melanotic encapsulation and results in sequestration and killing of foreign invaders in a blackened, melanin-derived capsule that effectively limits the number of parasites developing to the infective stage (Kobayashi et al. 1986; Beerntsen et al. 1989). This host–parasite system has emerged as an important model for characterization of the biochemical and molecular events comprising this form of invertebrate immunity (Zhao et al. 1995; Cho et al. 1997; Liu et al. 1997); however, virtually nothing is known of the genome organization of this mosquito host.
The linkage map presented herein provides the genetic context required for further examination of the genetic basis of melanotic encapsulation. Linkage maps based on common cDNA markers also allow for the testing of hypotheses concerning orthologous quantitave trait loci (QTL) associated with vector competence (Severson et al. 1995a). Use of linkage map-based technology has resulted in the identification of mosquito QTL that influence vector competence for nematode andPlasmodium parasites (Severson et al. 1994b, 1995b; Zheng et al. 1997).
RESULTS
Marker Evaluation
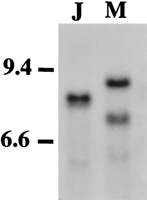
Fifty-six Ae. aegypti markers were screened on bulked,EcoRI-digested, DNA from the J and M Ar. subalbatusstrains, and 43 hybridized as heterologous probes (Table1). Strain-specific RFLPs were observed for the majority of these hybridizing probes, and 23 of these markers showed no common restriction fragments between the strains (Table 1). An autoradiograph of one of these informative probes, LF198, is shown in Figure 1.
Utility of Ae. aegypti Probes for Comparative Mapping with Ar. subalbatus
Autoradiograph of genomic blot probed with LF198 to screen for unique RFLPs between pooled DNA samples representing mosquito strains. DNA was digested with EcoRI. (Lane 1) Japan strain; (lane2) Malaysian strain. Size markers as determined by usingHindIII-digested λ phage DNA markers are given.
Segregation of RFLPs
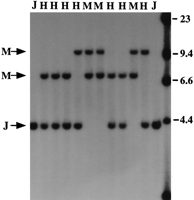
Twenty-six Ae. aegypti markers were probed to F2 progeny and scored with respect to the parental RFLP as either homozygous for the J or M genotype or heterozygous. Figure 2provides an example of an F2 segregating population probed with the chromosome 2 marker, LF115. Significant deviations from the expected 1:2:1 ratio were observed with 42% of the loci examined in at least one of the test crosses (Table2). Table 3 shows the observed segregation relative to sex, for the eight linkage group 3 RFLP loci. When these data were examined without respect to sex, most of these markers fitted the expected 1:2:1 ratio (Table 2).
RFLP observed in 10 segregating Ar. subalbatus F2progeny following hybridization with the Ae. aegypti LF115 probe. RFLP alleles representing the parental Japan strain (J) and Malaysian strain (M) are indicated. (H) Heterozygote. This locus is on chromosome 2. Size markers as determined usingHindIII-digested λ phage DNA markers are given.
Segregation of Markers in the F2 from Five Crosses Used to Construct a RFLP Linkage Map
Segregation of Linkage Group 3 RFLP Markers Relative to Sex
Linkage Analysis
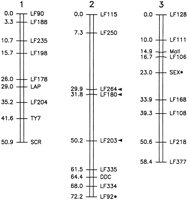
A linkage map was constructed by hybridizing DNA from segregatingAr. subalbatus individuals with 26 prescreened Ae. aegypti clones (Fig. 3). Linkage and linear relationships of markers on group 2 represent the best fit of data from four different crosses as determined by the JoinMap computer program (Stam 1993). Comparative information for the arrangement of these markers with respect to Ae. aegypti is a result of compilation of all available data. One marker, LF92, was mapped to a linkage group not predicted by the linkage arrangement observed for Aedes.Also, the sex determination locus, which maps to chromosome 1 inAe. aegypti, is located on group 3 in this Armigerescross. Finally, the linear positions of LF264, LF180, and LF203 on chromosome 2 are inverted with respect to the other group 2 markers as compared with Ae. aegypti. The loci mapped in Ar. subalbatus cover 181.5 cM, whereas the same markers in Ae. aegypti cover 181.3 (D.W. Severson, unpubl.), although the spatial distribution varies among linkage groups between these two species.
Genetic linkage map of Ar. subalbatus based on RFLP loci derived from Ae. aegypti. Map distances are given in centiMorgans. Asterisks denote markers that deviate from the linkage group predicted by the Ae. aegypti map. Arrowheads indicate three loci segment that is inverted with respect to its position onAe. aegypti linkage group 2.
DISCUSSION
Ae. aegypti cDNA markers hybridize under stringent conditions and exhibit differences in restriction fragment lengths between the two strains of Ar. subalbatus used in this study. Of 56 (77%) Ae. aegypti cDNA probes, 43 hybridized toAr. subalbatus DNA, confirming the high degree of gene conservation and usefulness of heterologous mapping between different species of culicine mosquitoes (Severson et al. 1994a, 1995a). The genetic diversity between the Armigeres strains for these markers (Table 1) is extensive and comparable to that reported forAe. aegypti markers used for mapping with Ae. albopictus, suggesting that there is relatively low residual heterozygosity between these two strains.
Segregation data for most RFLP loci fitted the expected Mendelian ratio (Table 2); however, there were several significant deviations. Related studies that produced a comparative map for Ae. albopictusresulted in significant χ2 values for 78% of the loci examined (Severson et al. 1995b), a result attributed to an abundance of the heterozygous genotype. Deviations likely reflect the lethal effects of loci on F2 progeny homozygous for the parental genotypes (Matthews and Craig 1989; Munstermann 1994). For the crosses presented here, this effect is most pronounced for markers on one end of chromosome 2, distal to LF115 (Fig. 3; Table 2), which exhibited a preponderance of heterozygotes and paternal (J) homozygotes in three of the four crosses used to study linkage on this group.
Sex determination in the Culicinae is inherited in a manner consistent with a single autosomal gene, with maleness being the dominant allele (Gilchrist and Haldane 1947). Males are heterozygous at the sex locus and females are homozygous for the recessive genotype, ensuring that homozygous males at the sex locus will not exist. When analyzed according to sex, it is obvious that the segregation pattern does not fit a 1:2:1 ratio (Table 3), particularly when approaching the sex locus. This result is consistent with a systematic segregation distorter on linkage group 3. The deviation should disappear when data are analyzed without respect to sex (Table 2), assuming an equal number of males and females are examined. The chromosome 3 marker deviations from 1:2:1 can be attributed to an inadvertent bias toward males in production of the mapping blots.
Genetic linkage and gene order is well conserved between these two species; however, several differences were observed (Fig. 3). Chromosomal repatterning has been studied in mosquitoes, and it was proposed that variation in linkage and linear order among mosquito groups can often be accounted for by translocations and inversions. In this case, tightly linked genes are more likely to share a chromosomal history; consequently, these genes “travel” together (Matthews and Munstermann 1994). The three central markers in Ar. subalbatus (LF264, LF180, and LF203) that do not map in the same linear order predicted by Ae. aegypti are inverted with respect to this species. One marker, LF92, maps to chromosome 2 inArmigeres and group 3 in Aedes. Database analysis of LF92 indicates a homology with the ubiquitin gene (D.W. Severson, unpubl.). Hybridization of this probe with Armigeres DNA is extensive, with a high degree of background signal. The only discernible polymorphism was scored. It is possible that this probe is detecting a family of genes, at several loci throughout the genome. A marker 3 cM away from LF92 in Ae. aegypti, LF111, occurs in the same linear order in both species (i.e., it has not been translocated with LF92).
The sex-determination locus maps to linkage group 1 in Ae. aegypti and group 3 in Ar. subalbatus, and variation in the sex-determination locus is strain-specific in another culicine genus, Culex. Geographic isolates of Culex pipiens quinquefasciatus vary in the position of this locus (Sakai et al. 1980). In another species, Culex tritaeniorhynchus, the sex locus segregates with chromosome 1 markers in one strain and chromosome 3 markers in another (Baker and Sakai 1976). The heredity of sex in culicine mosquitoes differs from that observed for Drosophila(MacDougall et al. 1995); however, it is unlikely the genetic mechanisms of sex determination and differentiation are entirely different between these two dipterans. It is likely that rather than vast differences in gene positions among species and strains of mosquitoes, genetic control is allelically based. From work inDrosophila it is clear that the molecular genetics of sex determination is a complex process requiring expression and interaction of many genes, and genetic varients might exist that transfer control of this cascade to a different gene in the pathway (MacDougall et al. 1995; Wilkins 1995).
The loci mapped in Ar. subalbatus cover 181.5 cM as compared to 181.3 cM for Ae. aegytpi. This congruency does not reflect the variation in recombinational distances according to linkage group, with Armigeres markers undergoing crossovers less on linkage group 1 and more on group 2 than their Aedes counterparts. The haploid genome size of Ar. subalbatus has been estimated as containing 1.24 pg of DNA (Rao and Rai 1990) as compared to 0.81 pg forAe. aegypti (Rao and Rai 1987). It is not clear why this difference in nulear DNA content is not reflected in recombination frequencies, but recent data suggest the genome size for Ae. aegypti is significantly smaller than that reported by Rao and Rai (1990); similarly, it is possible that the size of the Ar. subalbatus genome also is smaller (D. Zaitlan, pers. comm.).
Comparative mapping can be a powerful approach to genetically characterize a previously undescribed organism, and generation of genetic linkage maps recently has provided the necessary tool to gain insight into the heritibility of mosquito susceptibililty to several parasite species (Severson et al. 1994b, 1995b; Zheng et al. 1997). TheAr. subalbatus genome is now defined in discrete units that can be used to identify non-random association of genomic regions with vector competence using QTL procedures in conjunction with crosses between parasite-resistant and -susceptible line of Ar. subalbatus. As data from related studies accumulate, comparative QTL studies also might elucidate common inheritance patterns associated with vector competence and suggest novel approaches for the control of vector-transmitted diseases.
METHODS
Mosquito Strains and Crosses
Two Ar. subalbatus strains were used for these studies. One strain originated from Japan (J), and was provided by G.B. Craig (University of Notre Dame, South Bend IN), and the other strain was colonized in Malaysia (M) and was provided by Dr. Akio Mori (Institute for Medical Research, Kuala Lumpur). The RFLP linkage map was based on five segregating F2 populations resulting from sibling matings of F1 populations. The F1 populations were generated from a mass mating between J males and M females.
Restriction Length Fragment Polymorphism Markers
All of the RFLP markers used in this study are Ae. aegypti cDNA clones with the exception of Scr (Sex-combs-reduced), which is a probe derived from the homeobox region of theDrosophila melanogaster gene (Mahaffey and Kaufmann 1987) recently mapped in the Ae. aegypti genome (D.W. Severson, pers. comm.). Markers were selected from throughout the Ae. aegypti genome, with the goal of obtaining similarly complete coverage of the Ar. subalbatus genome. Most of these clones represent random cDNAs (Severson et al. 1993). MalI andLAP represent known Ae. aegypti cDNAs—salivary gland maltase (James et al. 1989) and lysosomal aspartic protease (Cho and Raikhel 1992), respectively.
DNA Isolation, Southern Blotting, and Hybridization
DNA extracted from pools of mosquitoes representing each of the parental (J and M) strains of Ar. subalbatus and from F2 individuals, was EcoRI-digested, Southern blotted, and hybridized according to methods described previously (Severson et al. 1993; Severson 1997). Briefly, total nucleic acids were isolated by homogenization in equal volumes of lysis buffer and phenol, followed by standard phenol/chloroform extraction (Sambrook et al. 1989). This isolate was incubated with RNase A followed by a second extraction with phenol/chloroform. Ae. aegypti markers initially were used to screen bulked DNA preps from each of the parental strains to verify hybridization and to identify informative loci. Ten-microgram aliquots of mosquito genomic DNA were digested withEcoRI, size-fractionated on 0.9% agarose gels, and blotted to Hybond (Amersham) membranes. F2 mapping blots were prepared similarly using DNA from single individuals. Presceening identified informative RFLP markers (Table 1) from which 26 were chosen for hybridization with (J × M) F2 progeny for generation of mapping data. Probe insert preparation and radiolabeling were accomplished using a PCR protocol with defined primer annealing sites of the vector plasmid flanking the mosquito DNA (Severson 1997). Prehybridization, hybridization, and membrane washing procedures were conducted at 65°C in glass bottles in a commercial hybridization oven (Hybaid) (Severson et al. 1993; Severson 1997). Membranes were washed using stringent conditions (twice in 2× SSC with 0.1% NaDodSO4 for 15 min, and twice in 0.2× SSC with 0.1% NaDodSO4 for 15 min) before exposure to film for 3–6 days at −80°C with an intensifying screen.
RFLP and Linkage Analysis
χ2 goodness-of-fit ratios were determined for segregation and independent assortment of alleles for all pairs of loci. Multipoint linkage analyses were performed using Mapmaker software (Lander et al. 1987) with a minimum lod threshold of 3 used to identify nonrandom association between markers. The Mapmaker program is unable to compile data generated from different crosses; consequently, a composite linkage map was developed for chromosome 2 and for inclusion of the sex determination locus on group 3, using the JoinMap computer program (Stam 1993). The Ar. subalbatus linkage map was depicted graphically using DrawMap (van Ooijen 1994). Map distances are reported as centiMorgans (Kosambi 1944).
Acknowledgments
We thank L. Christensen and J. Klinkhammer for assistance in mosquito rearing, and V. Kassner and L. Beaty for their participation in blot preparation and hybridization. This work was supported by National Institutes of Health grant A1-19769 (to B.M.C.) and the Burroughs Wellcome Fund (to B.M.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
-
↵1 Corresponding author.
-
E-MAIL bmc{at}ahabs.wisc.edu; FAX (608) 262-7420.
-
- Received September 18, 1997.
- Accepted December 2, 1997.
- Cold Spring Harbor Laboratory Press