WebWise: Guide to The Sanger Center’s Web Site
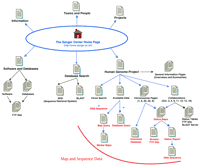
This installment of the WebWise series reviews The Sanger Center’s web site (http://www.sanger.ac.uk/). The Sanger Center, one of the larger sequencing centers involved in the Human Genome Project, has made the necessary commitment to establish and maintain a world class web site. This web site also has a uniform style, which has the twofold benefit of making the whole site feel like a cohesive package, as well as enhancing the browsing experience. The Sanger Center provides a wealth of data on its web site; at the time of this writing, it reports having finished >30 million bases of human DNA sequence. Their main sequencing efforts are chromosomes 1, 6, 20, 22, and X, but they are also collaborating on sequencing small regions of chromosomes 3, 4, 9, 11, 12, 13, and 16. The site map depicted in Figure 1 illustrates the overall organization of this web site, and the main features of the web site are highlighted in Table1. This web site is extensively interlinked, and for some features several alternate routes are provided to access the data; for example, links to the FTP site are provided on several different pages. In general, navigation is not difficult because a convenient header is provided at the top of most pages that allows the user to maneuver between different areas of the web site. Note that these internal links, as well as most duplicate or minor links, have been omitted from Figure 1.
The Sanger Center Site Map. The main links to pages discussed in the text are illustrated here. Links on the top of the Home Page are to general informational resources; the links located on thebottom portion are to the data or additional tools.
Features of The Sanger Center Web Site
General Information
The links located at the top of Figure 1 (Information, Teams and People, and Projects) lead to general information related to the genome sequencing effort. Contact information is provided via the Home page link to Teams and People and also through the Search option located directly under this link. Information concerning other genome projects being carried out at the Sanger Center is available by following the Projects link. Following the Information link leads to a page providing links to some general information about the Sanger Center, including travel and job information. A table summarizing the amount of finished and unfinished sequence data generated is available by following the Progress Statistics link, which is also provided at the bottom of the Home page. A useful list of relevant links is also accessible from the main Information page (http://www.sanger.ac.uk/Info/Links) and includes hot links to numerous genome centers, model organism databases, other biological databases and resources, and some commercial sites. Although this is a fairly comprehensive list, it was noted that a few genome centers and bioinformatics resources are not included in the pertinent list.
Data
Because many interpage links are provided on this web site, the discussion below represents, in many instances, only one of several routes to a given web page. To access the human genome data being generated at this site, follow the Human Genome Project link from the Home page. The resulting page (http://www.sanger.ac.uk/HGP/) consists of an organized series of links to Sanger Center resources and data, including overviews and summaries, search engines, collaborations, and chromosome-specific data. It is easy to miss a very useful search tool, provided at the bottom of this page, that allows you to search for the sequence of a given clone. Upon searching for a clone (dJ130G2 was picked randomly from the chromosome 6 status map) a status summary result is returned with a link to the FTP site FASTA sequence provided at the bottom of the page. Of course, you must already have a clone name at hand, but once this information has been obtained (by browsing the maps or FTP site) this tool is a convenient method to continue to monitor sequencing progress of a given clone.
Information about the larger sequencing targets on chromosomes 1, 6, 20, 22, and X is organized by chromosome, with links to chromosome-specific web pages provided from the Human Genome Project page. Follow the Collaborations link to learn more about the smaller collaborative projects on chromosomes 3, 4, 9, 11, 12, 13, and 16. Although some chromosome-specific information is provided for the major sequencing efforts, the chromosome-specific pages are organized similarly. A link to the chromosome status map is provided toward the top of the page, and following this link leads you to an image map of the chromosome. You can zoom in to higher resolution maps by successively clicking on the region of interest. For example, after following the link to the Chromosome 22 status map you can zoom in on the 22q12.1 region by clicking on the boxed regions to the right or left of this text. You can follow this “intermediate” resolution view to the high-resolution map by clicking on the red rectangles. Alternatively, you go directly to the high-resolution map from the first image by clicking on the red regions. Take care to click within the rectangle regions; clicking on a blank area above or below a rectangle yields variable results, including occasionally calling up a map page containing only a broken link symbol. The high-resolution maps provide an indication of the sequence status (color-coded rectangles on the left), the clone tiling path, and the clone name (listed on the right side of the display). It is very convenient to navigate from the higher resolution map data to the sequence data by simply clicking on the clone name or status rectangle. This brings up a status report reviewing the progress for the selected clone and provides a link directly to the DNA sequence on the FTP site. If you wish to navigate through the FTP site, follow the link to the Human Sequence directory of the FTP site that is provided above the image maps (ftp://www.sanger.ac.uk/pub/human/sequences). The status maps, generated using ACeDB [A C. elegans Database (Thierry-Mieg and Durbin 1991)], as well as the sequences on the FTP site, are updated nightly. Although it is very easy to navigate through the maps, interpreting the status is hampered by a broken link to the map legend figure.
Additional chromosome-specific information is available by following the links below the Status map link. On individual chromosome pages, the links under Mapping and Sequencing provide a description of the methodology and progress. Most of these pages include a Data Available/Mapping/Genomic Sequence set of links that provide further descriptions and links to some very useful search tools, such as Chromosome-specific Database Query Pages and Query for Maps (see the Tools section below). A second map view is accessible with one of these search tools; once you know a contig or clone name that is located in a region of interest, you may find it useful to try the Query for Maps feature (http://www.sanger.ac.uk/HGP/db_query/search_for_map.shtml). The maps (Marker Maps, Fig. 1) displayed with this search engine differ slightly from the status maps by integrating a variety of data and markers including sequence status, STS (sequence-tagged site) markers, clones, and named loci. Although this is not an interactive image map, further information about the clones and markers can be obtained by utilizing the search tool provided at the bottom of the page. Although this is a very useful feature, it is not convenient to navigate to and from these pages, as you must navigate through a few pages before you can use the map search tool, and the next several layers of web pages do not include links back out to other pages on this site. For instance, you cannot simply click on a link to return to the Home page or Human Genome page but must instead “go back” several pages. Furthermore, links are not provided to the DNA sequence or FTP site from these map views; nonetheless, these integrated map displays should be quite useful for some tasks, including positional cloning.
Tools
In addition to providing a wealth of map and sequence data, this web site provides several valuable resources, including (1) several search tools, (2) the means to carry out BLAST searches against The Sanger Center databases, (3) downloadable chromosome databases, and (4) descriptions of the software and databases used by, and available at, The Sanger Center. Together, these features expand the overall utility of this web site immensely by making it convenient to find data, compare your sequences to The Center’s data, and download maps, sequences, databases, and programs.
The Sanger Center is obviously committed to making its data accessible as it supplies the necessary tools to facilitate data retrieval via its web site. Tools are provided to search for a clone’s sequence as well as for both a sequence status map and a map that integrates sequence status and marker data. These tools are extremely useful and are accessible from several web pages, including the chromosome-specific pages (http://www.sanger.ac.uk/HGP/Chr#/; follow the Data Available link; the most direct route is to follow the Data Available link from the Human Genome Project page). This brings up a page that includes links to three different search options but does not include either the option to search for a clone’s sequence or to download a chromosome-specific database. Searching for a clone’s sequence is only available from the bottom of the Human Genome Project page (http://www.sanger.ac.uk/HGP/), and downloading a database is discussed below. To find a higher resolution sequence status map you can either browse the lower resolution map of the relevant chromosome or utilize the map query tool (follow the Query for Maps link from the Data Available page;http://www.sanger.ac.uk/HGP/db_query/search_for_map.shtml). You can search for maps using either a Marker or Clone name. Once you have identified a particular map that you are interested in (and make note of a marker or clone name), this tool provides a convenient mechanism to monitor sequencing progress of that region over time. One further search tool is the option to query the chromosome-specific databases. This page (http://www.sanger.ac.uk/HGP/db_query/query.shtml) can be reached from the Human Genome Project page by following first the Data Available link, and then the Chromosome Specific Databases Query link. The preformatted Queries page provides more search options than the map search tool and enables the user to search for data by key word, gene name, or map features. The preformatted Queries map search option allows the user to search for a single marker by type (e.g., STS) or for all markers between two positions. Although some explanatory text is available directly from the Preformatted Query page, a better explanation of the options is available at a less accessible page (http://www.sanger.ac.uk/HGP/db_query/query_help.shtml#keyword). For those who wish to have full query access to the databases, an Open Access button will let you type in your query directly using Tace syntax; help documentation on the Tace query syntax is provided.
One additional search service, the Sequence Retrieval Service (SRS), is also available on The Sanger Center’s web site. This service, which is also available at many additional sites around the world, allows the user to search the available molecular biology databases using a single search engine. Although, in theory, this should save the user a considerable amount of time by eliminating the need to search each database individually, in practice this service is complex and difficult to implement. Although advanced documentation is provided in the form of an online manual, beginner-level documentation to outline the general use or provide some examples is not available.
The Sanger Center also makes available many software tools and provides extensive documentation on these tools (http://www.sanger.ac.uk/Software/). Although these software tools are really too numerous to describe thoroughly here, they include programs relevant to generating maps (Image 3.x, FPC, SAM), sequencing (Common Assembly Format, sequencing software), and sequence analysis (GCG Extensions, WiseTools, Dotter, Blixem). Overall, the documentation is quite useful and includes a general overview of the programs as well as instructions for downloading, installing, and using the programs. Additional information on the ACeDB database and the Pfam database (a collection of protein domain families and alignment data) is also accessible from this page.
One unique feature of The Sanger Center’s web site is the ability to download the chromosome-specific ACeDB databases. Several of the internal chromosome-specific pages include a link and directions for downloading and installing these databases. Unfortunately, the links to this supportive documentation are inconsistent, as are the links to the FTP site. To reach a description of this service you must jump down a series of links (e.g., Human Genome Project to Chromosome 1 to Available Chromosome 1 Data to Search our Available Chromosome 1 Data) or go directly to http://www.sanger.ac.uk/HGP/Chr1/datare.shtml. This page presents a description of the computer requirements and software you will need to download. An alternative route to the chromosome database FTP sites is available via the Software page. Follow the link to ACeDB (http://www.sanger.ac.uk/Software/Acedb/) to access documentation on ACeDB as well as more convenient links to the chromosome database FTP sites (seeftp://ftp.sanger.ac.uk/pub/human/chr#/). Additional instructions for downloading the databases are provided in README files on the FTP site.
The Sanger Center web site also includes a Human BLAST server that allows you to search their sequence data for a sequence of interest. This is an extremely useful tool provided to the research community: One simply pastes the sequence of interest, in FASTA format, into a text area box (or use the Browse option to select a file on your computer). Unfortunately, this BLAST server is not as flexible as desired, as results are only returned by e-mail, sometimes up to several days after posting the inquiry. An empty e-mail message can also be the result of your search (as was the situation in one of our test cases), leaving one wondering whether an error occurred or whether there were no significant matches—it would benefit the user to have a simple statement to the effect that no homologous sequences were identified. In general, this service has the potential to be extremely useful to the user, but, as it stands, it could be enhanced. At a minimum, it would be nice to have the option to display the BLAST result directly on the browser window, preferably with links provided to any homologous sequences.
Conclusions
The Sanger Center’s web site provides a significant amount of map and sequence data, as well as numerous useful support services, including search tools, software, and databases. The web site has, for the most part, adopted a simple uniform style that has the overall effect of facilitating navigation. A few of the innermost pages do not include the useful navigation header and would benefit from an upgrade. The map and sequence data are easy to locate, and several different search tools are available that facilitate locating a particular map or sequence. More importantly, The Sanger Center has successfully integrated the map and sequence data by providing “clickable” image maps for the status maps and linking them to the sequence data.
Some of the information that is relevant to a single tool is provided on multiple pages that are then accessed from different routes. When these pages are combined with the overall extensive interlinking of the web pages, navigation of the inner pages of this site can be somewhat confusing, especially for the first-time user. For example, the chromosome-specific web pages do not always use consistent nomenclature or provide consistent links to supportive documentation and tools, so it is not immediately clear, for instance, if a given link will lead you to text documentation about mapping or to a map-related search tool. It is a challenging task to structure and organize a large web site, and The Sanger Center has achieved this with the top layer pages; however, the organization of some of the inner pages could be improved.
Although a very useful assortment of search tools are provided, the tools are not all easily accessed. Given the number of search options available at this site, it seems reasonable to expect a single page that provides a comprehensive series of links to the different search tools and downloadable data. This single page should be directly accessible from the Home page and “linked to” from the navigational header as well as from the body of pertinent pages. The Data Available page, accessible from the Human Genome Project page, is an unsuccessful attempt to fully integrate these resources. For instance, the Data Available page does not include the capability to search for a clone or to download a chromosome database. Furthermore, the chromosome-specific web pages do not link back to the Data Available page but, rather, provide variable link options either directly to the search engine or to additional text pages that include the link to the search tools. This inconsistent approach does impede efficient navigation of the web site. Those who are interested in performing searches or database downloads on a routine basis must navigate through the web site or bookmark the relevant page(s). These points aside, one must give credit to The Sanger Center for making the necessary commitment to maintain a web site that does provide a significant amount of data and other important resources to the research community. This is a very large web site and requires a committed ongoing effort to maintain it; The Sanger Center does a better job than most at maintaining a unified style and providing consistent navigation links.
Next Month: The Stanford Human Genome Center and the Whitehead Institute/MIT Genome Sequencing Project.