Closing in on Telomeric Closure
The mapping of STS markers to subtelomeric chromosome regions is complicated by unique features of subtelomeric DNA. Low-copy repeat elements are very abundant in subtelomeric regions and thus cause problems in generating robust, locus-specific STSs. An STS priming site in a repeat sequence alters the effective ratios of primers in the PCR reaction (making the assay less reliable), and the two STS priming sites in a low-copy repeat result in the generation of an STS that maps to multiple genomic sites (sometimes on the same chromosome). YAC-based STS content map information is extremely limited in subtelomeric chromosome regions because YAC clones carrying this DNA are under-represented in most YAC libraries. Conventional YACs derived from subtelomeric regions are often prone to rearrangement. Finally, because the positioning of STS markers at the ends of maps is constrained only by mapping data from one side of the marker, there is a tendency for “problem” markers with genotyping ambiguity or error to be positioned at the ends of maps rather than within maps (where their position is more rigorously constrained by mapping data from flanking markers on both sides).
In sum, robust, single-copy STS-based markers for distal subtelomeric regions are difficult to find; subtelomeric marker orders based on genetic linkage, radiation hybrid (RH), and STS content mapping methods are more error-prone than normal, and clone coverage of most subtelomeric regions in YAC libraries is sparse. If subtelomeric regions were not so interesting, it might be wise to avoid them entirely. However, subtelomeric regions are interesting. They have enhanced levels of recombination, very high gene densities, and their sequences may contribute to and be modulated by the telomere dynamics associated with telomeric (TTAGGG)n tract length variation (Greider 1996) and epigenetic alterations of telomeric chromatin and heterochromatin.
Enhanced recombination frequencies and expansion of genetic linkage map distances relative to physical map distances are common features of human subtelomeric regions, particularly for male meioses (Rasmussen and Holm 1978; Donis-Keller et al. 1987; Rouyer et al. 1990; Blouin et al. 1995). This means that higher than normal genetic marker densities are required for coverage of a given physical human subtelomere region in linkage studies. In the absence of terminal markers physically linked to chromosome ends, this also means that an indeterminate (and potentially large) genetic distance might exist between the most distal mapped genetic marker and the actual telomere. The DNA in this subtelomeric interval might not be adequately represented in genetic screens for linkage of genome regions to disease phenotypes.
Subtelomeric DNA has very high gene densities (Saccone et al. 1993;Flint et al. 1997a), including genes and gene families whose evolution and function are likely to be influenced by the elevated levels of subtelomeric recombination (Cook et al. 1994; Kermouni et al. 1995;Martin-Gallardo et al. 1995; Amann et al. 1996; Van Deutekom et al. 1996). Novel recombination and/or transposition-based mechanisms of subtelomeric gene regulation (Navarro and Cross 1996; McCulloch et al. 1997) and telomere maintenance (Levis et al. 1993; Lundblad and Blackburn 1993; McEachern and Blackburn 1996) have evolved in widely divergent eukaryotic genomes, and telomere-specific chromatin and heterochromatin-like structures are believed to influence the expression of subtelomeric genes in yeast and in Drosophila(Gottschling et al. 1990; Weiler and Wakimoto 1996; Donaldson and Karpen 1997). Recent data indicate that recombination-based pathways of telomere maintenence exist in human cells (Murnane et al. 1994; Bryan et al. 1995; Rogan et al. 1995) and are characterized by very long and heterogenous telomeres. The functions of subtelomeric DNA sequences might be modulated by the dynamic aspects of telomeric chromatin structure and by the variation of telomeric (TTAGGG)n repeat tract lengths in cells. Finally, large-scale chromosome length polymorphisms have been detected at many human telomeres (Wilkie et al. 1991; Ijdo et al. 1992; Martin-Gallardo et al. 1995), and their causes, frequencies, and potential functions are not understood. Telomeric closure of STS maps is an essential step toward a comprehensive analysis of subtelomeric sequence features and is necessary for evaluating and characterizing these putative functions of subtelomeric DNA.
Now, Rosenberg et al. (this issue) describe the isolation and mapping of STS markers for the distal-most single-copy regions of 28 human chromosome arms (see Fig. 1 for examples). Given the probable absence of unique sequences on the p-arms of the five acrocentric chromosomes, this means that STS-based maps of 28 of the 41 unique human chromosome arms now have firm telomeric boundaries. A set of cloned fragments that included half-YACs, subtelomeric P1 clones, and telomeric cosmid contig clones (used previously as subtelomere-specific FISH probes; Ning et al. 1996) was used as source material for these markers. The half-YAC clone libraries (Riethman et al. 1989; Kvaloy 1993) provided cloned telomere–terminal fragments of human chromosomes. Validation of close physical linkage to telomeres had been demonstrated conclusively (in previous work) for a sizable fraction of these clones, and compelling (but not conclusive) evidence suggests that most or all of the remaining half-YAC clones are also very closely linked physically to their respective telomeres. Thus, one can be relatively confident that the markers of Rosenberg et al. (this issue) represent the distal-most single-copy chromosomal regions and can be used to close the telomeric gaps in the genome map.
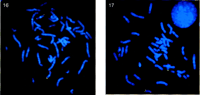
FISH using probes specific to telomeres from chromosome 16 and 17, as indicated. The p-arms are labeled with red and the q-arms are labeled with green. (The probes and technique used are as described in Knight et al. 1997).
The immediate utility of the new markers is illustrated by the mapping results described in Rosenberg et al. (this issue). At some telomeres, the marker order is perfectly consistent with expectations, with the new markers mapping to the ends of the linkage groups. However, at several telomeres the data demonstrate discrepancies between map orders using the new telomere markers and the expectation that the new markers should be at the ends of the respective linkage groups. These subtelomeric regions merit more detailed analysis, as unexpected marker orders might reflect large chromosome-length variations as well as possible genotyping errors in the database. One chromosome-length variation may be a 50-kb polymorphism at 2-qter (Macina et al. 1994); a marker for this telomere appeared to be hemizygous in some CEPH families (Rosenberg et al., this issue).
As expected from previous studies, analyses of the sequences used in generating the subtelomeric markers revealed segments of DNA sequence similarity between different subtelomeric regions. This work was extended by Flint and coworkers recently and revealed an intriguing pattern of subtelomeric repeat features. Most striking was the presence of a degenerate internal TTAGGGn repeat adjacent to a consensus sequence for the putative eukaryotic origin of replication; this pair of motifs was present in the human subtelomeric regions examined and at all of the 32 Saccharomyces cerevisiaesubtelomeric regions [in spite of the fact that the S. cerevisiae telomerase adds (TG1–3)n repeats rather than (TTAGGG)n repeats (Flint et al. 1997b)]. It was proposed that the adjacent internal (TTAGGG)n and putative replication origin motifs demarcate a boundary between distal and proximal subtelomeric chromosome compartments, each of which is postulated to have separate chromatin structures and functional properties (Flint et al. 1997b).
What of the remaining telomeres? Ongoing work has resulted in the identification of cloned half-YAC fragments derived from each of the remaining nonacrocentric telomeres (with the exception of 5p; Ning et al. 1996; H.C. Riethman, unpubl.). Discrete STSs have already been prepared from each of these clones (with the exception of clones believed to be derived from the 12p and 20q telomeres, for which there are presently no single-copy STSs available; H.C. Riethman, unpubl.). Once these additional STSs are placed on the RH maps, telomeric closure for a human genome map will be nearly complete.