Rare Disease Genes—Lessons and Challenges
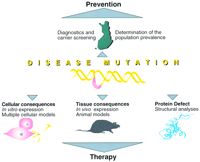
The fiercest competition in the field of human genetics takes place in the area of genetic diseases that are common at the population level—this is primarily attributable to the potential commercial utilization of emerging data. However, dissection of the molecular background of many extremely rare diseases has proven to be highly useful for the detailed characterization of cellular dysfunction and the identification of novel metabolic pathways, broadening our understanding of biological processes in general. This information has largely been obtained from research carried out in populations with exceptional enrichment of a given disease (Fig. 1).
The identification of a disease mutation is considered beneficial, as it provides tools for assessing the pathogenic mechanisms of the disease and ultimately designing prevention and therapy. Research into the molecular background of many rare disorders has also provided the scientific community with various new strategies for the locus identification of a disease gene, as well as given new insight into metabolic pathways and biological processes.
Although the global prevalence of rare diseases is insignificant [e.g., worldwide there are <200 known cases of patients with infantile-type neuronal ceroid lipofuscinosis (INCL) in comparison to a global population prevalence of 1:65 for the recessive cystic fibrosis mutation or 1:8000 for dominant Marfan syndrome], this rarity does not necessarily reflect the impact of these diseases on biological research.
From Population Sample to Gene Identification: Special Statistical Strategies
Rare diseases are characteristically enriched in populations that have been isolated for religious, linguistic or geographical reasons; good examples of this are the Ashkenazi Jews or the Finns. From the viewpoint of population genetics, rare diseases have initially led to the identification of many population bottlenecks and have produced quite precise data on the time period at which the mutation was actually introduced into the population, thus providing initial clues to the genetic history of human populations. Often a common ancestor or inbreeding can be verified in the history of affected individuals, which suggests that there is one major ancestor mutation. This situation allows the adoption of unique strategies to search for a causative locus: similarity searches for the disease locus using DNA samples from just a few affected individuals and searches for shared alleles or homozygosity of genotypes. Using this approach, an initial genome scan with 400 markers can be carried out in just a few days as compared to the weeks or months required for genotyping all family members in traditional genome scans using linkage analysis in diseases that have a more diverse mutational background. The strategy of using only a few DNA samples has been highly successful in the locus identification of many rare diseases (Nikali et al. 1995; van Soest et al. 1996 , this issue).
The phenomenon of shared alleles in affected individuals or significant deviation in the allelic frequencies of polymorphic markers in disease chromosomes compared to control chromosomes is called linkage disequilibrium. This is sometimes observed with markers over amazingly wide intervals, as exemplified in Table 1, which summarizes the data on Finnish disease alleles. The existence of linkage disequilibrium over such broad intervals suggests that genome-wide association-based analyses—perhaps not well-justified in mixed, heterogeneous populations—could also be successful for locus searches in complex polygenic diseases in genetic isolates (Lander and Schork 1994).
Interval Showing the Linkage Disequilibrium in the Disease Alleles of Finnish Disease Heritage
The linkage disequilibrium-based strategy has also been utilized successfully to restrict the critical DNA region after initial assignment of the disease locus. This is perhaps best exemplified in the isolation of the gene defective in a severe cartilage disorder called diastrophic dysplasia. Utilizing the rules of bacterial genetics (Luria and Delbrück 1943) and a simplified population genetic model based on the assumption of a small number of ancestors in the Finnish population, the gene was originally predicted to be located 64 kb from the polymorphic marker showing the strongest linkage disequilibrium. When the gene, which encodes a sulfate transporter protein, was isolated, it was 70 kb from this marker (Hästbacka et al. 1994). Again, applications of this method for complex diseases and their predisposing genes are obvious. Characteristically the DNA region identified to be associated with a common trait is very wide because of statistical problems combined with the incomplete penetrance of hypothetical disease gene(s). Monitoring of linkage disequilibrium in population isolates ought to be a highly powerful tool for the more precise localization of predisposing genes. However, the situation in complex diseases is likely to be more problematic than in the case of rare disease alleles because of the expected high population prevalence and multiplicity of predisposing mutations—even in an isolate.
Shortcuts to Gene Functions and Clues to Essential Biological Pathways
Well-characterized tissue symptoms in patients with rare diseases provide immediate understanding of the function and tissue expression of the identified disease gene. On many occasions, totally new metabolic pathways have either been identified or associated for the first time with a specific cell type or developmental stage. Again, some Finnish diseases exemplify this: The identification of the mutated gene for diastrophic dysplasia provided evidence of the exceptional sensitivity of cartilage cells to the relative lack of sulfate ions as compared to other cell types (Hästbacka et al. 1994). Similarly, the finding that the causative gene for the lethal infantile brain disorder INCL encoded a palmitoyl protein thioesterase demonstrated for the first time that proper removal of palmitoyl residues from lipid-modified proteins is an absolute requirement for the normal development and maturation of neocortical neurons (Vesa et al. 1995). INCL is characterized by the rapid death of cortical neurons, whereas neurons in lower parts of the CNS remain intact. Although details of both diastrophic dysplasia and INCL remain to be characterized further, these findings have opened new avenues for research of the metabolism of both sulfate and palmitoyl residues in specific cell types and tissues.
A lysosomal enzyme deficiency, aspartylglucosaminuria (AGU), resulting in progressive mental retardation, serves as a good example of a very rare disease that might initially be regarded as rather uninteresting. However, years of intensive research into the pathogenesis of AGU exposed molecular details of more general relevance. At first, the need for elucidation of the disease mechanism stimulated research into the defective enzyme, aspartylglucosaminidase (AGA), which had so far been hampered by failure to purify the protein to sufficient homogeneity because of its low quantity in tissues. Following identification of the gene and the major disease-causing mutation, in vitro expression studies of normal and mutated AGA polypeptides clarified the details of its biosynthesis and intracellular processing and provided clues to some general features of the cell biology of lysosomal enzymes (Ikonen et al. 1991a,b, 1993; Mononen et al. 1993; Riikonen et al. 1994, 1996). Subsequent crystallization data on the AGA enzyme revealed a novel catalytic mechanism based on the amino-terminal nucleophile and indicated that AGA is the first eukaryotic member of a new enzyme family of amidohydrolases (Oinonen et al. 1995; Tikkanen et al. 1996a,b). Structural analyses also provided the means for characterizing the lysosomal targeting process of AGA in detail and increased our comprehension of the targeting of lysosomal enzymes in general (R. Tikkanen, M. Peltola, C. Oinonen, J. Rouvinen, and L. Peltonen, in prep.).
From a purely biological standpoint, the study of rare diseases has potential similar to research with knockout mice, which have been used traditionally for characterizing the tissue consequences of a gene defect. The phenotype of these rare diseases is extremely well described, and despite the complexity and individual variability of the genetic background, skillful clinicians have typically confirmed the unifying clinical features between patients. Consequently, once the mutation has been identified, the function of a defective gene can be dissected immediately at a tissue- or cell lineage-specific level. Moreover, well-monitored follow-ups of these patients over many years produce detailed information on the disease process—and all information is directly applicable to humans! When the monitoring of individual variations in multiple genes becomes more feasible, for example, by array technology, even higher accuracy will be possible in the perception of the tissue-specific consequences of single genes mutated in human diseases.
The examples presented here for the enormous potential that comes from the molecular characterization of rare diseases were chosen from one population. Similar information has emerged from numerous other rare disorders, the classical example being the low-density lipoprotein (LDL)–receptor mutation in familial hypercholesterolemia that revolutionized our concepts of lipid metabolism (Brown and Goldstein 1986). Within the next several years all human genes will be cloned. This development will provide scientists with more precise tools to tackle the basic molecular defects in all of the >4000 rare human diseases (McKusick 1994), the causes of which still remain unknown. The vast amount of information emerging from this endeavor will have immense significance for the basic concepts of the multiple metabolic routes in human cells and tissues.