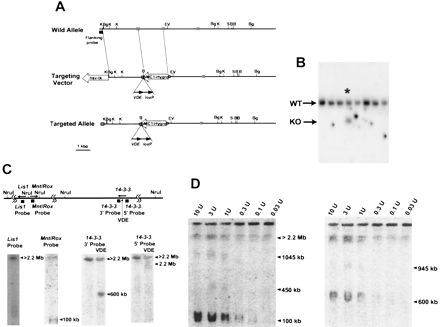
14-3-3ε targeting of VDE site for determination of transcriptional orientation. (A) The restriction map and the targeting vector are shown. pMC1hyg and PGK–tkindicate the genes used for positive and negative selection with hygromycin and FIAU; arrow directions indicate transcriptional orientation. VDE and loxP sites are indicated. The dotted boxes indicate 14-3-3ε exons. Restriction sites are as follows: (B) BamHI; (Bg) BglII; (EV)EcoRV; (H) HindIII; (K) KpnI; (S)SalI; (VDE) the super-rare cutter. (B) The result of the screening of the transfected ES cell by Southern hybridization using a flanking probe and BamHI digestion. (Asterisk) The positive clone; (WT) wild-type allele; (KO) knock-out allele. (C, top) The NruI restriction map of the MDS region in the mouse, with the location of the three genes (the direction of transcription indicated by arrows), and the location of probes used for hybridization. (Bottom) PFGE and Southern hybridization of DNA from the targeted ES clone with a VDE site inserted into the14-3-3ε locus. DNA was digested with NruI alone (nothing above the lane), or NruI and VDE (indicated by VDE above the lane), and hybridized with the indicated probes. The arrows indicate the sizes of the bands detected with each probe. (D) PFGE Southern hybridization for complete VDEdigestion and complete or partial NruI digestion. The enzyme concentration per microliter is above each lane. The arrowheads indicated the band detected by the 14-3-3ε 3′ probe.











