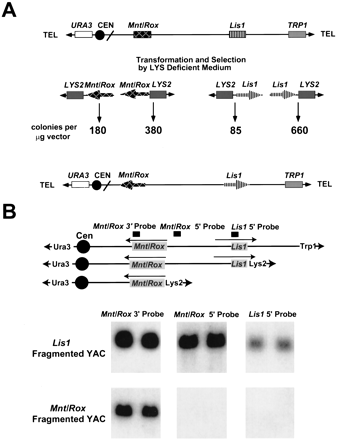
Lis1-Mnt/Rox YAC fragmentation for transcriptional orientation. (A) Schematic representation of YAC fragmentation. (Top) The Lis1-Mnt/Rox YAC clone is shown, with the location of the two genes, URA3 andTRP1 selectable markers, and the position of the centromere and telomeres. (Middle) The fragmentation vectors withLYS as the selectable marker near the telomere, and theMnt/Rox (left) or Lis1 (right) cloned in both orientations relative to the telomere. Underneath the fragmentation vectors, the number of colonies per microgram of vector are indicated. (Bottom) The gene orientation on the YAC relative to centromere was inferred by comparison of the recombinant frequency of each fragmentation vector. The arrows indicate the 5′ → 3′ direction of each gene. (B) Southern analysis of the fragmented YAC. (Top) The original YAC is shown on the first line, followed by the organization of theLis1- and Mnt/Rox-deleted YACs. The locations and transcriptional directions of the two genes, as determined by colony number, and the locations of three hybridization probes are shown. The arrows indicate the 5′ → 3′ direction of each gene. (Bottom) DNA from two LYS+ transformants from theLis1 fragmentation, and two from the Mnt/Roxfragmentation, were digested with EcoRI, transferred to nylon membranes, and probed with three different probes: a Mnt/Rox3′ probe, a Mnt/Rox 5′ probe, and a Lis15′ probe.











