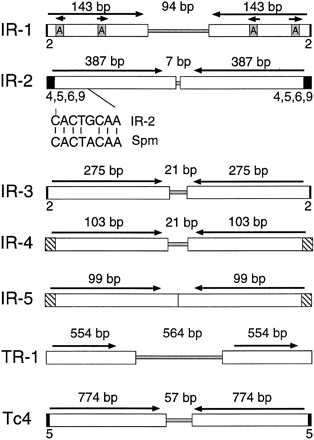
Structures of the IR and TR repetitive elements. The IR and TR elements are shown, indicating the presence of inverted or tandem repeats (white boxes), spacer regions (shaded regions in the center), and flanking sequence duplications (small black boxes). The terminal sequences of IR-2 and Spm are compared under the IR-2 map, with 7/8 base pair identity. The hatched areas of IR-4 and IR-5 indicate their similar terminal sequences. The Tc4 transposon of C. elegans, which has been shown to be a functionally active mobile element in C. elegans (Yuan et al. 1991), is shown for comparison. Note that only full-length representative copies are shown for each repetitive family and that each family was as structurally diverse as IR-1 (Fig. 4).











